
Immune System Development
Human Immune System Development Map (HIDmap)
Online access and exploration: https://immunedev.elixir-luxembourg.org/minerva/
Development status: Fist version is complete and published
Sustainable support: Environmental Toxicants and the Brain, MINERVA Platform
License: Creative Commons Attribution 4.0 International (CC BY 4.0) License
Construction tool: CellDesigner
Funding: Clariant Produkte (Deutschland) GmbH and IUF - Leibniz Research Institute for Environmental Medicine
Contact: Christiane Spruck, IUF – Leibniz Research Institute for Environmental Medicine, christiane.spruck(at)iuf-duesseldorf.de
Description
The development of the human immune system is a biologically complex process involving multiple cellular niches and diverse cell types across distinct developmental stages. Current knowledge in this field largely relies on animal studies, highlighting the need for in vitro and in silico new approach methodologies (NAMs) that enable the assessment of effects on the developing immune system without animals use.
To capture the biological complexity of human immune system development and to support the design of such NAMs, we have initiated the construction of the Human Immune System Development Map (HIDmap). This physiological map (PM) – a concept derived from the Disease Maps project - focuses on the representation of normal, undisturbed physiological processes. Based on extensive literature curation, the HIDmap provides a graphical overview of the key cellular niches and intricate biological interactions occurring during prenatal immune development, including cell migration, cell-cell interactions, and signaling molecules. The major anatomical sites involved comprise the aorta-gonad-mesonephros (AGM) region, yolk sac, fetal liver, bone marrow, spleen, and thymus. Cell types represented in the HIDmap include, but are not limited to, hematopoietic stem cells (HSCs), progenitor cells, T-cells, B-cells, dendritic cells, macrophages, and monocytes.
The main goal of the HIDmap is to comprehensively map human immune system development to identify knowledge gaps and provide a valuable reference for both research and regulatory applications in pharmacology and toxicology. In this context, the HIDmap aims to serve as a foundation for the development of adverse outcome pathways (AOPs), computational models, and human-relevant in vitro NAMs for toxicological testing - ultimately supporting the establishment of a NAM-based test battery for developmental immunotoxicity (DIT).
Publications
Spruck et al., 2025 (submitted)
HIDmap User Guide
This quick guide explains how to navigate and explore the HIDmap.
Accessing the HIDmap
The main entry point is: https://immunedev.elixir-luxembourg.org/minerva/. Upon opening, you will see an overview diagram displaying all organs included in the map, along with three buttons on the left-hand side.
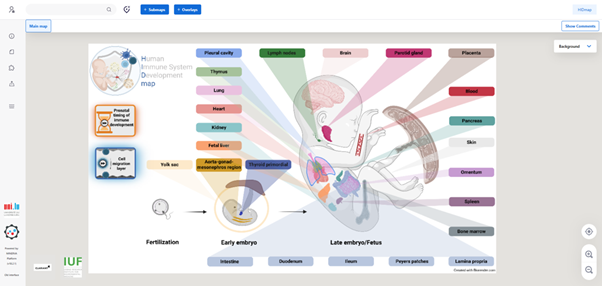
Navigating Organ Maps
To view a detailed maps for a specific organ, click on its name in the overview diagram (red arrow).
For example, selecting ‘Fetal liver’ opens the corresponding submap.
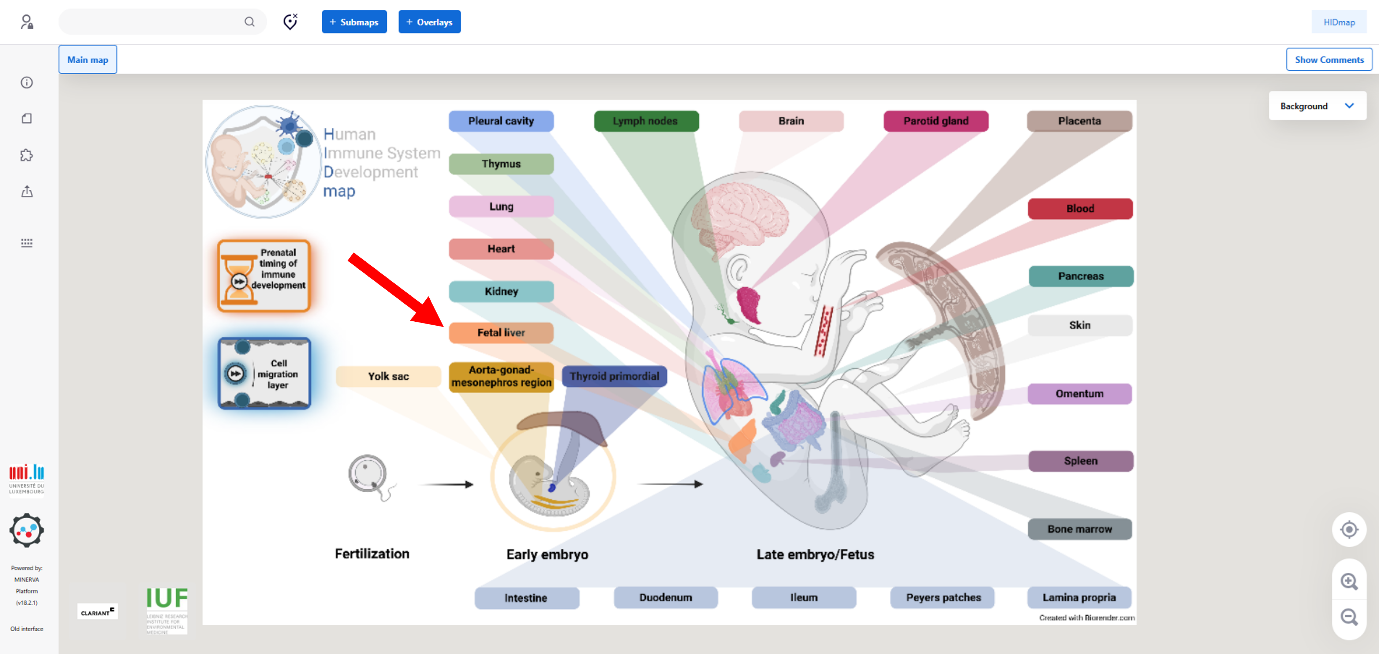
On the left-hand side, three clickable images provide shortcuts to:
• The Disease Maps homepage (HIDmap logo; top button; red arrow)
• The prenatal development of the human immune system overview (middle button; orange arrow)
• The cells migration map, showing inter-organ connections (bottom button; green arrow).
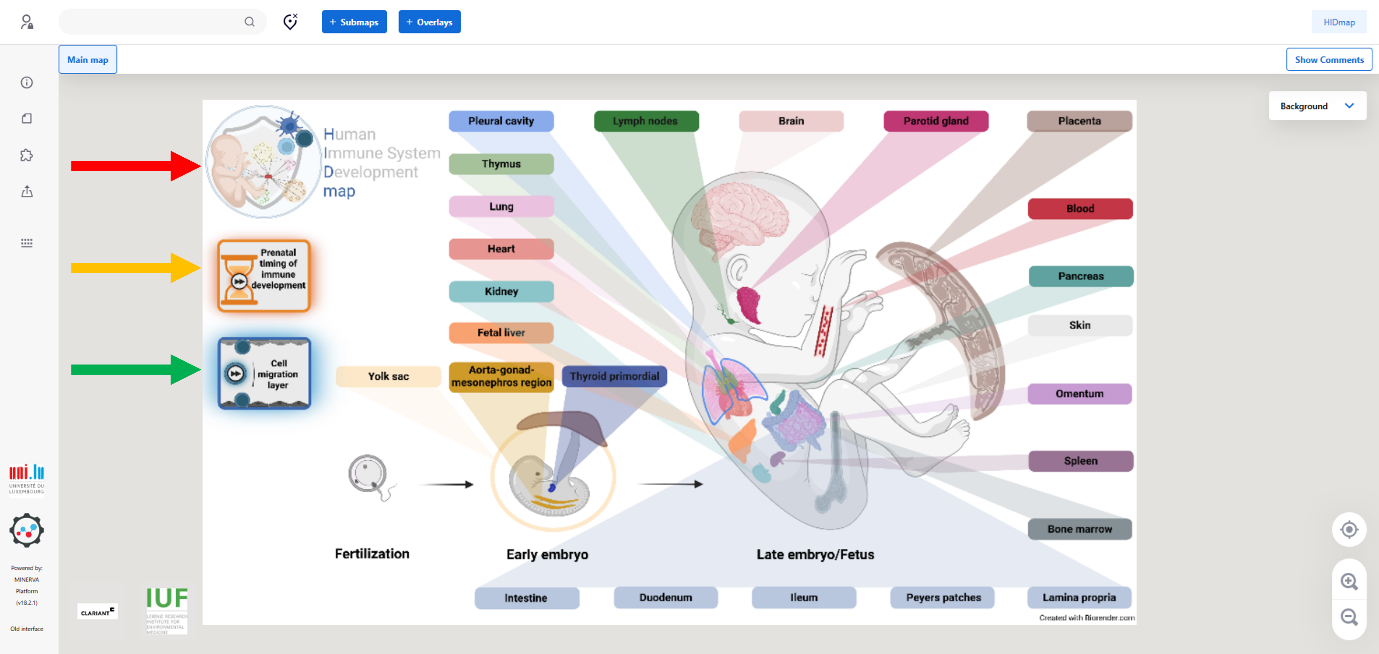
Pan and zoom
Navigation within the HIDmap works similar to Google Maps:
• Use the buttons on the bottom right (see the three arrows) to zoom in, zoom out, and re-center the view.
• Click and drag to move around the map.
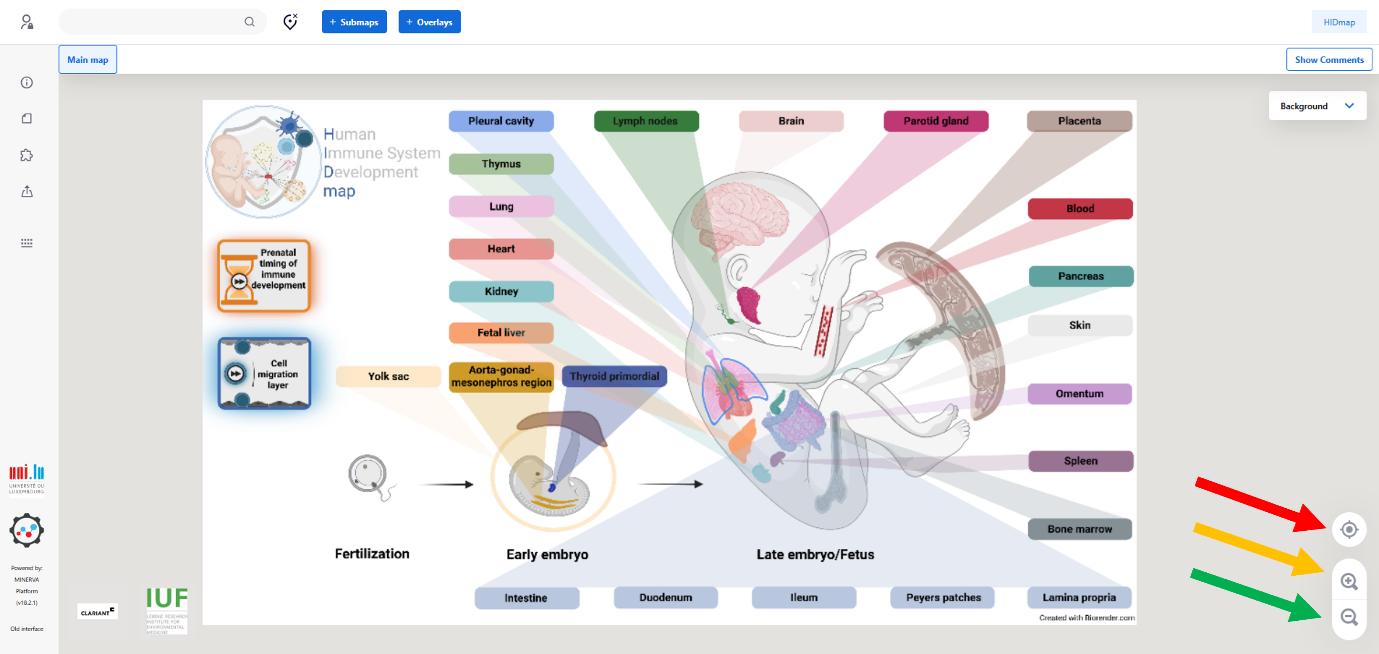
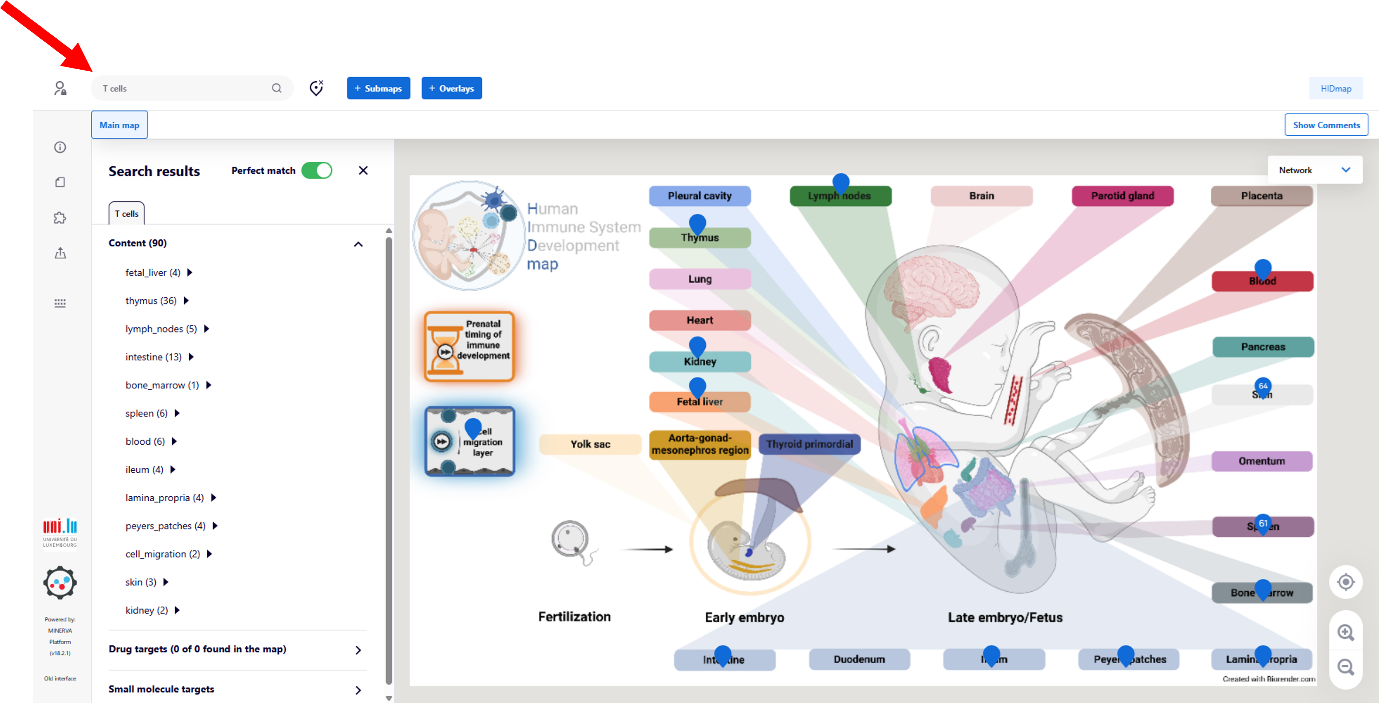
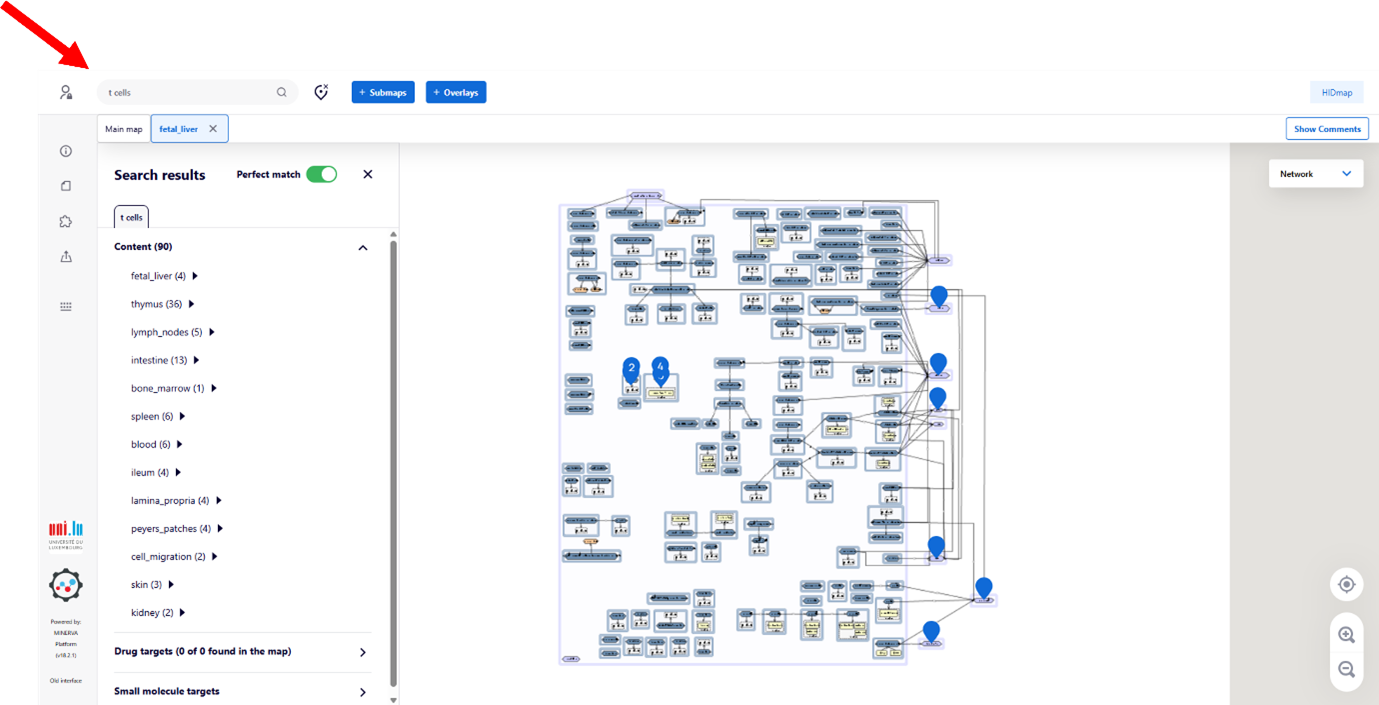
Search Function
You can search for specific elements using the search field (red arrow at the top left). Results are highlightedon the map with blue anchors. To clear the search, click the ‘delete’ button.
Search functionality is available both in the overview figure and within each submap.
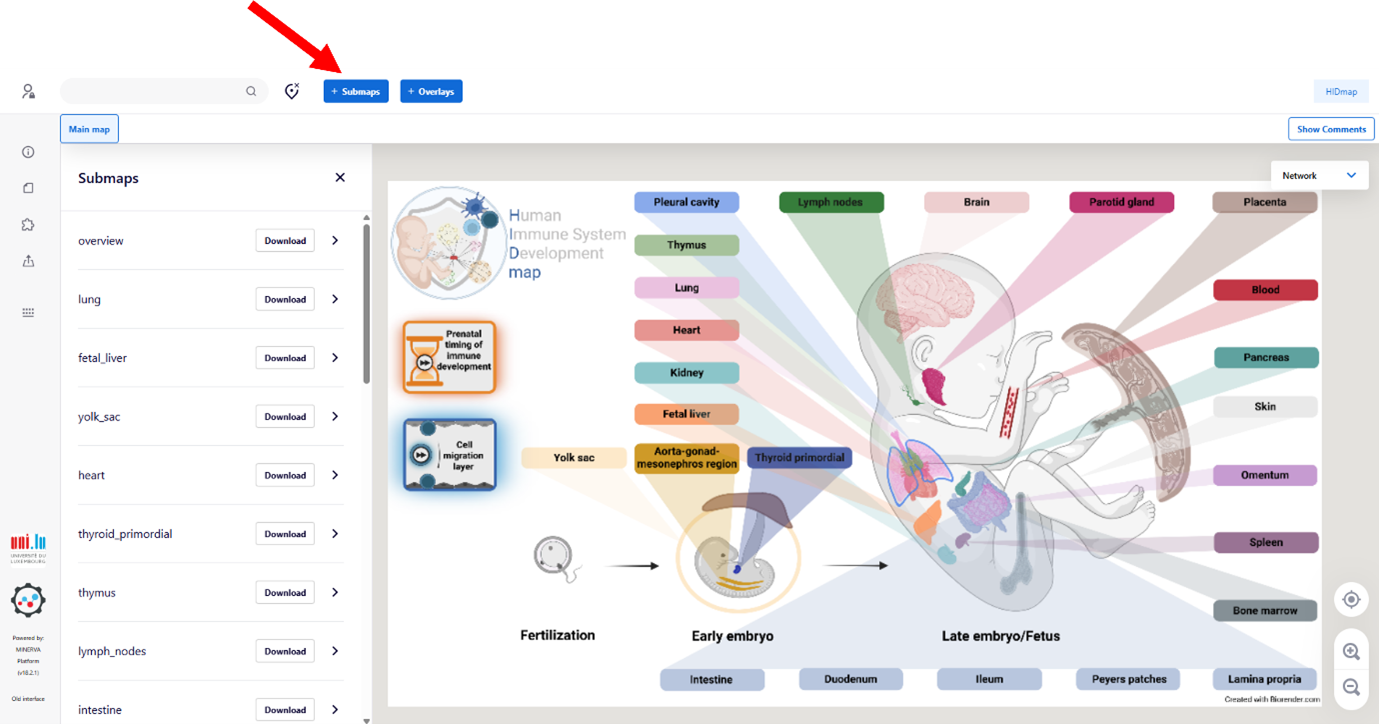
Submaps
To explore submaps, click on the ‘Submaps’ button at the top (red arrow). A will appear in the left-hand panel, allowing access to:
• The main submaps (e.g. Cell Migration and the Prenatal Immune Development)
• Organ-specific submaps (e.g., Fetal liver, Thymus, Bone Marrow, etc.)
Interactive exploration
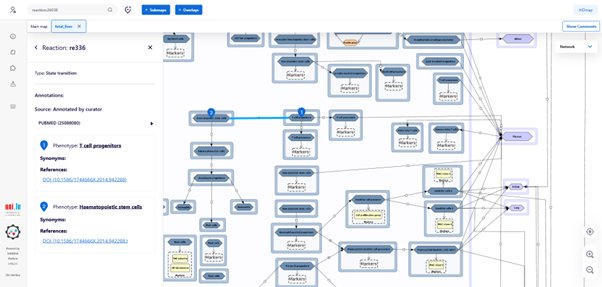
Every map element – such as cells, markers, or interactions - is clickable. Selecting an element opens a detailed annotation on the left-hand side, which provides:
• Curated biological information
• Official HGNC gene symbols
• Supporting PubMed references.
All molecular markers are systematically annotated with official HGNC symbols (A). Non-standard markers (e.g., major histocompatibility complex (MHC) class II) appearing yellow boxes (B), ensuring clear differentiation and traceability.
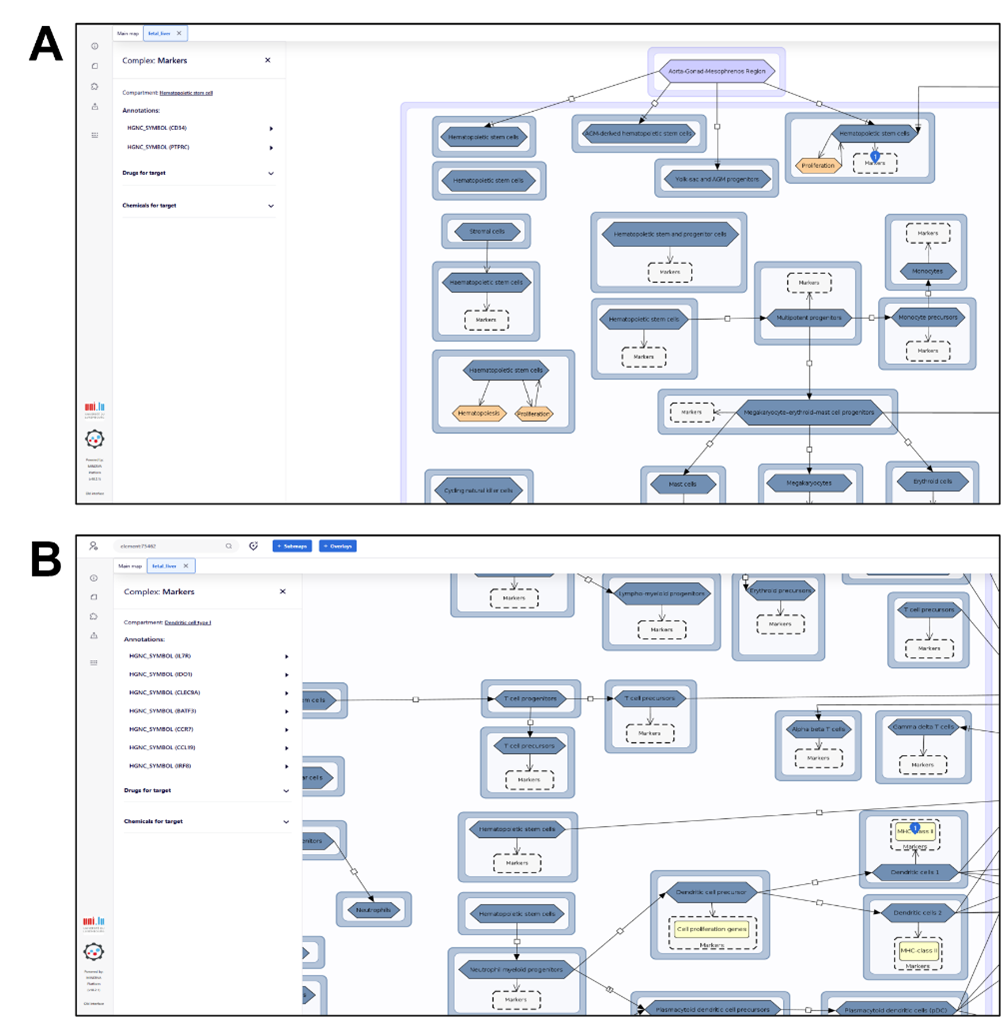
Clicking on an interaction (black connecting lines) displays the relevant publications supporting that biological relationship.
Tip
Use the combination of search, submaps, and interaction details to explore the developmental pathways and inter-organ relationships of the developing human immune system.
Development Team
 |
Christiane Spruck, M.Sc IUF – Leibniz Research Institute for Environmental Medicine Biologist, PhD Student Development of the HIDmap |
 |
Luiz Ladeira, PhD GIGA Institute, University of Liège, Belgium Postdoctoral researcher Curation of the HIDmap |
 |
Eliška Kuchovská, PhD IUF – Leibniz Research Institute for Environmental Medicine Postdoctoral researcher Review and editing |
 |
Marek Ostaszewski, PhD University of Luxembourg, Luxembourg Luxembourg Centre for Systems Biomedicine Researcher MINERVA Platform support |
 |
Emanuela Corsini, PhD DISFeB – Università degli Studi di Milano Professor of Toxicology Scientific Advisor |
 |
Liesbet Geris, PhD GIGA Institute, University of Liège, Belgium Professor in Biomechanics and Computational Tissue Engineering Supervision |
 |
Bernard Staumont, PhD GIGA Institute, University of Liège, Belgium Postdoc Researcher & Project Manager Supervision, review and editing |
 |
Susann Fayyaz, PhD Clariant Produkte (Deutschland) GmbH Expert Toxicology Project administration and funding body |
 |
Qiang Li, PhD Clariant Produkte (Deutschland) GmbH Senior Expert Toxicology Project administration and funding body |
 |
Fabian Grimm, PhD Clariant Produkte (Deutschland) GmbH Head of Toxicology and Ecotoxicology Project administration and funding body |
 |
Ellen Fritsche, MD Swiss Centre for Applied Human Toxicology (SCAHT), Basel, Switzerland Director IUF – Leibniz Research Institute for Environmental Medicine Former working group leader Medical Faculty, Heinrich Heine University Düsseldorf, Duesseldorf, Germany DNTOX GmbH, Duesseldorf, Germany Professor for Environmental Toxicology Project generation, funding acquisition, administration and supervision |
 |
Katharina Koch, PhD IUF – Leibniz Research Institute for Environmental Medicine DNTOX GmbH, Duesseldorf, Germany Scientist and interim working group leader Supervision, project administration and funding acquisition |
 |
Julia Tigges, PhD IUF – Leibniz Research Institute for Environmental Medicine Scientist Supervision, project administration and funding acquisition |
Funding
This PhD project is funded by the company Clariant Produkte (Deutschland) GmbH and the IUF - Leibniz Research Institute for Environmental Medicine.
Acknowledgements

|
This project is supported by ELIXIR Luxembourg (ELIXIR-LU) Node. ELIXIR-LU hosts and maintains the MINERVA Platform for this project and supports its development. |