
Applications of the ISD map
Here we demonstrate the ISD map can be a hypothesis-generating resource via the discovery of potential mechanistic downstream effects of selected AD- or PsO-related genes of interest. For this purpose, we either analysed the network structure of the map itself or analysed the outcomes of omics data integration into the map.
Analysis of the network structure
Discovering possible pathways in KC that counteract the anti-inflammatory effect of dupilumab
Step-by-step guide to reproduce the analysis here
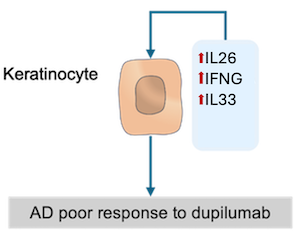
The direct analysis of the network structure per se may provide biological insights related to a disease of interest. For instance, to find pathways in KC that could attenuate dupilumab’s anti-inflammatory effect, we analysed the network structure of the KC submap in AD (KCs). We could identify pathways in KCs that could mitigate, at least partially, the anti-inflammatory effects of dupilumab: IL26, IL33 and IFNG signaling pathways. These pathways regulate CXCL8, TSLP and TNC, all of which are also regulated by the IL4/IL13 signaling pathway. So,the presence of IL26, IL33 and IFNG in skin could disrupt the effects of dupilumab.
Integration of genetic variation data into the map
We integrated AD- and PsO-related genetic variation data (obtained from the Open Targets database) and, after analyzing the positions of the affected genes in the map, more specifically their encoded proteins, we could formulate the hypotheses shown below.
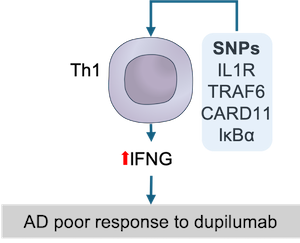
Data-driven hypothesis: SNPs in upstream IFNG regulators in Th1 cells favor upregulation of IFNG and drive resistance to dupilumab.
Step-by-step guide to reproduce the analysis here
We collected genes harbouring variants (SNPs) associated with AD from the Open Targets Genetics database and then mapped them to the ISD map (see these genes (in orange) in the map). After integration of these AD-associated genes to the map, we sough to investigate their influence at the mechanistic level. To this end, we manually inspected the pathways of the ISD map for proteins encoded by the matched disease-associated genes that directly influence other proteins. As discussed previously, IFNG seems to partially compensate for IL4R inhibition by positively stimulating the expression of several AD-promoting genes stimulated by IL4R in KCs. As IFNG is mainly produced by Th1 cells, we checked the Th1 cell map for the presence of proteins encoded by AD-associated genes that could somehow influence IFNG expression. Interestingly, there are five proteins encoded by AD-associated genes (IL18RAP, IL18R1, TRAF6, CARD11 and NFKBIA) upstream to the IFNG expression. We hypothesise that SNPs in these genes could favour IFNG expression in Th1 cells and, therefore, counteract the action of dupilumab, i.e., IL4R inhibition.
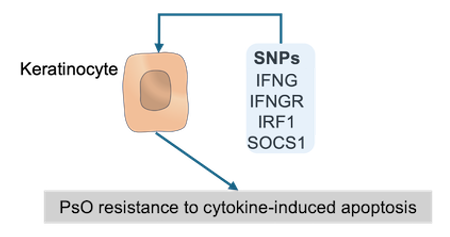
Data-driven hypothesis: SNPs in upstream apoptosis regulators in psoriatic KCs drive their resistance to cytokine-induced apoptosis.
Step-by-step guide to reproduce the analysis here
We collected genes harbouring variants (SNPs) associated with PsO from the Open Targets Genetics database and then mapped them to the ISD map (see these genes (in green) in the map). After integration of these PsO-associated genes to the map, we sough to investigate their influence at the mechanistic level, specifically in psoriatic KCs. In PsO, KCs are relatively resistant to cytokine-induced apoptosis. This resistance could be assigned, at least partially, to the presence of several proteins encoded by PsO-associated genes in apoptosis-regulating pathways. In fact, starting from keratinocyte apoptotic process, we can see a upstream pathway until the plasma membrane containing only proteins encoded by PsO-associated genes, namely, SOCS1, IRF1, IFGNR2 and IFNG. In this pathway, IFNG activates IFNGR2 that, in turn, stimulates the expression of IRF1. IRF1 then triggers the expression of SOCS1 that is known to inhibit apoptosis. Therefore, we can hypothesize that the underlying SNPs could favour the inhibition of apoptosis in KC (see figure below).
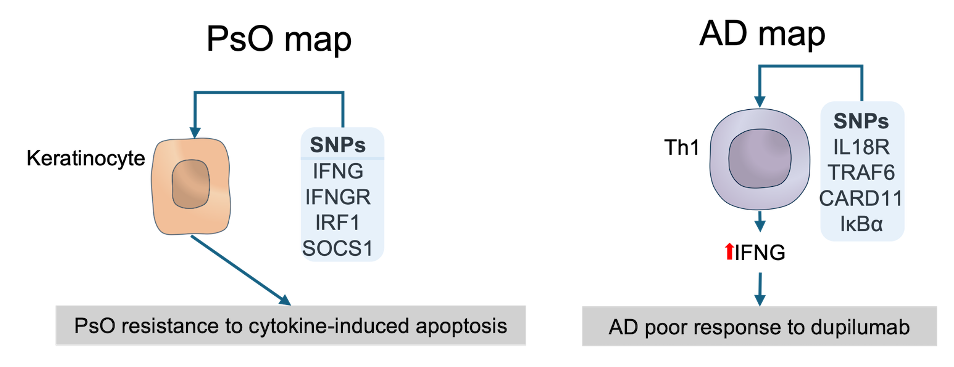
The above examples show that, through the integration of Open Target Genetics data with ISD map, we could determine the contextual relevance for AD- and PsO-associated genes and check if they fit into the existing molecular and cellular understanding of AD and PsO. Moreover, we were also able to formulate two hypotheses: resistance to dupilumab due to enhanced expression of IFNG in Th1 cells favoured by SNPs in upstream IFNG regulators and resistance to cytokine-induced apoptosis in psoriatic KCs due to altered upstream apoptosis regulators. The figure below depicts these hypotheses.
Integration of transcriptomics and proteomics data into the map
We integrated AD- and PsO-related omics data (obtained from biomedical literature and Gene Expression Omnibus database) and, after analyzing the positions of the differentially expressed genes and proteins in the map, we could formulate the hypotheses shown below.
Data-driven hypothesis: IL26 rescues the expression of TSLP, CCL20 and IL33 in AD keratinocytes under dupilumab treatment
Step-by-step guide to reproduce the analysis here
We collected differentially expressed proteins (DEPs) from the study by He et al. (2020) in which proteome expression profiles were measured in AD lesional and non-lesional stratum corneum samples taken from patients before and after treatment with dupilumab. From this study, we considered only DEPs calculated by comparing expression profiles of 353 inflammatory proteins extracted from lesional stratum corneum samples of patients before and after dupilumab exposure. Of the 132 dupilumab-induced differentially expressed inflammatory proteins (Dup-DEIPs), 20 could be found in the AD map (see these proteins in the map here).
We sought to check the KCs intracellular activity for finding which and how these Dup-DEIPs are distributed in KCs intracellular pathways. Of the 34 inflammatory proteins in KCs, nine (IL1RL1, IL17RA, IKBKB, CASP3, CASP8, CXCL8, CCL17, TNF and MMP9) are Dup-DEIPs, all being downregulated by dupilumab. We first checked which of these Dup-DEIPs are regulated by IL4/IL13 signalling. Of these nine proteins, only CXCL8 is a IL4/IL13 target and, as expected, it is downregulated. While CXCL8 is downregulated by dupilumab, the expression of TSLP, another IL4/IL13-induced inflammatory protein, seems not to be affected; TSLP is also regulated by IFNG signalling according to the map, so the TSLP expression could be rescued by IFNG in the absence of an active IL4R. IL17RA is downregulated by dupilumab and, therefore, we would also expect a downregulation of IL17RA signalling inflammatory target proteins. However, the DEP data provide no evidence that either of its inflammatory targets in KCs, namely CCL20, CSF3 and IL33, are affected by dupilumab. Among these proteins, it is possible to see that IL33, as TSLP, is also regulated by IFNG. So, IL33 expression could also be rescued by IFNG.