
Step-by-step guide to reproduce the application “Discovering possible pathways that counteract the anti-inflammatory effect of dupilumab”
1. Access and log in to the map
1.1. Access the ISD map at the entry level via the link (https://imi-biomap.elixir-luxembourg.org/). In the (map), click the login icon in the left upper side of the screen. This is required to integrate data into the map.
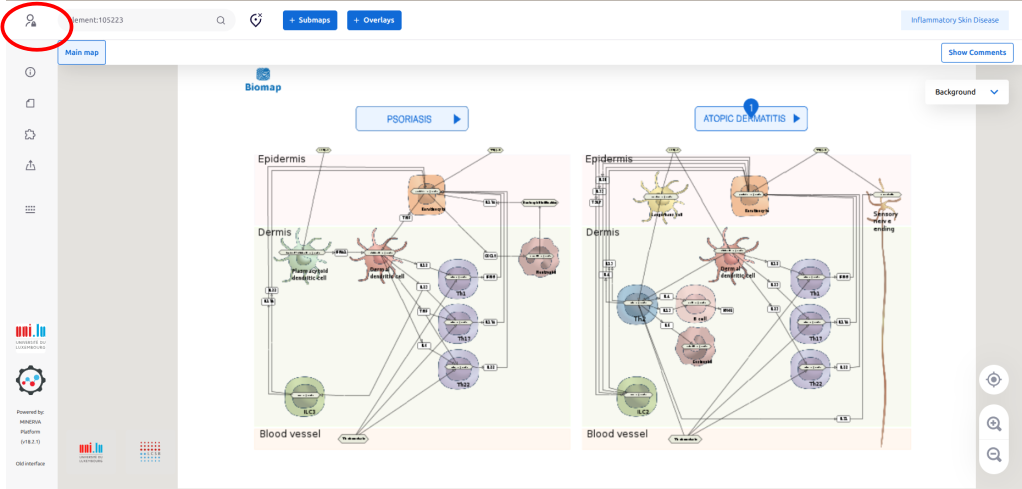
1.2. Log in to the map preferentially by using your ORCID.
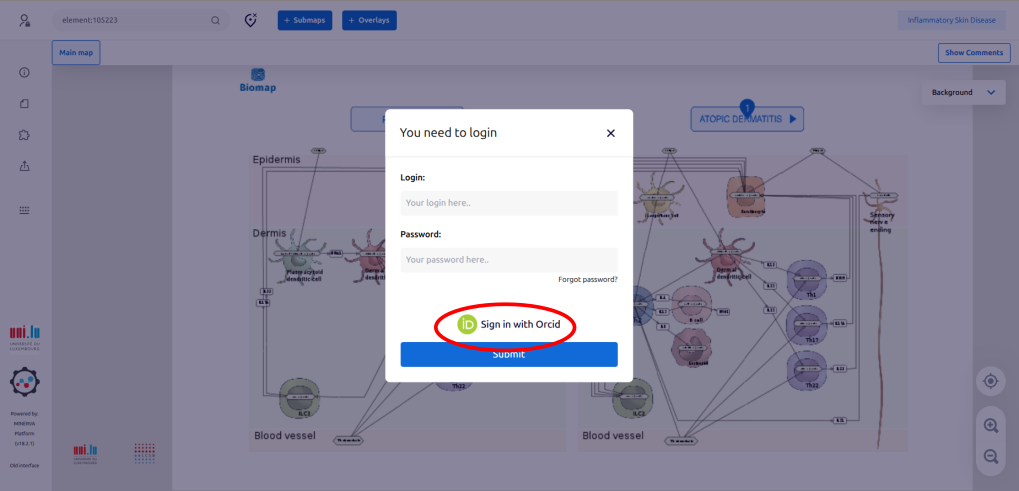
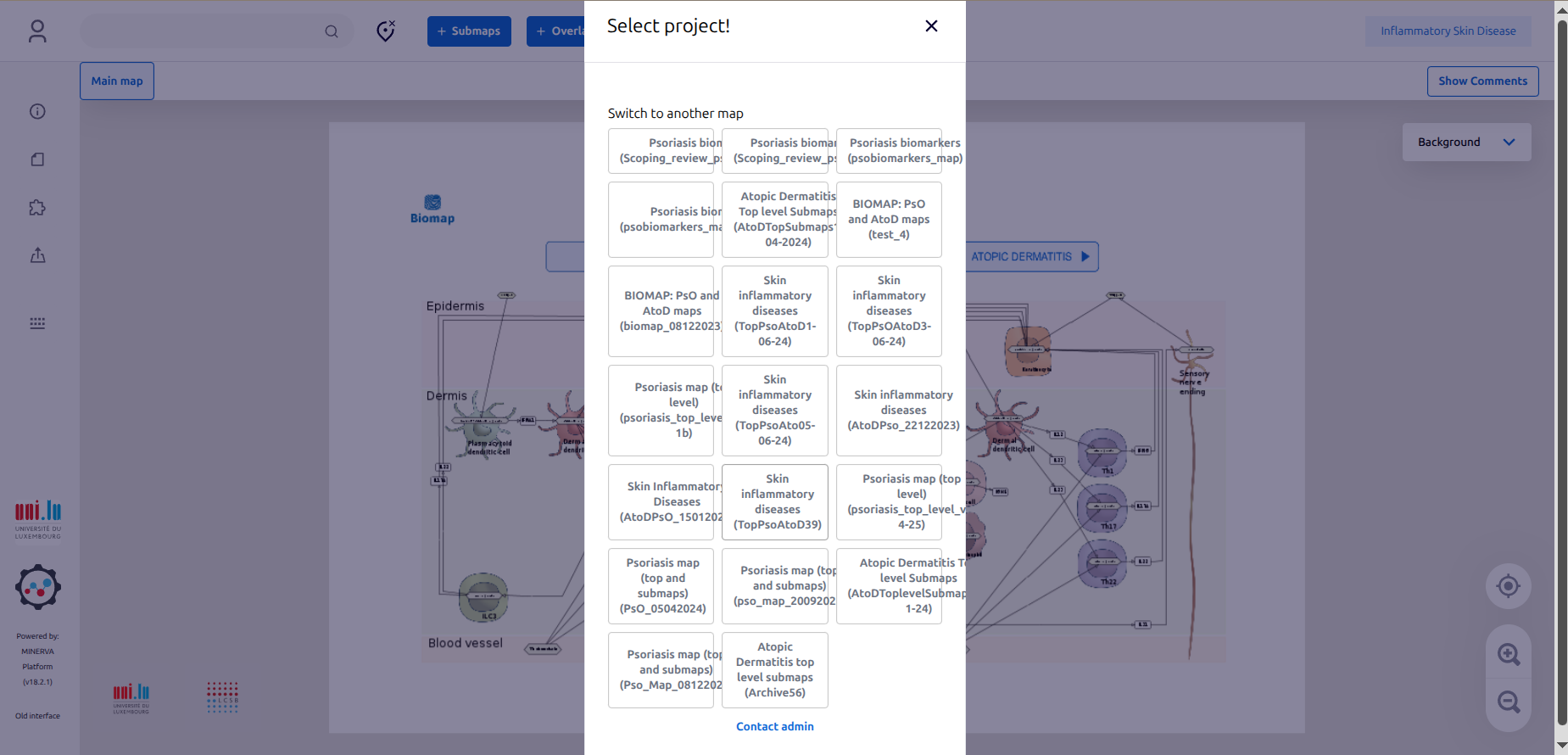
1.3. Once connected, just ignore the window “Select project” by clicking “X”.
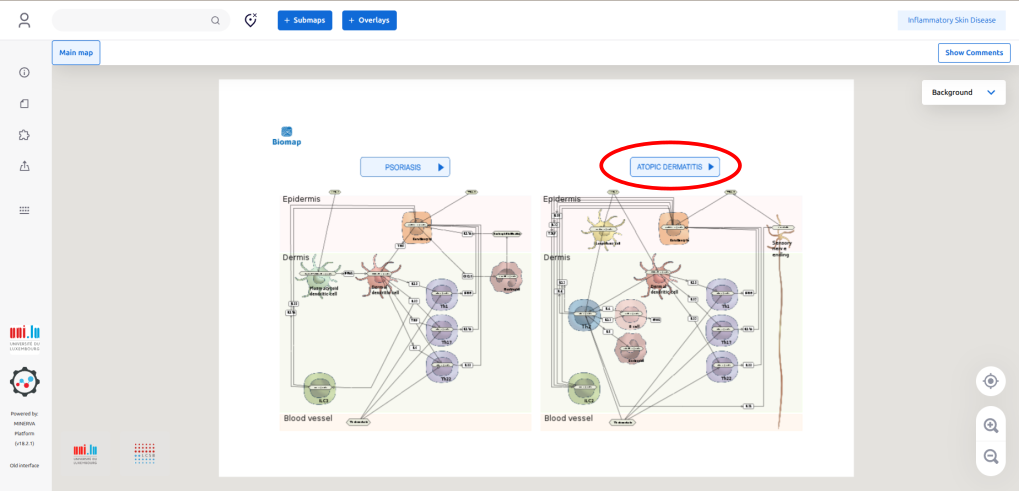
1.4. Click the button “ATOPIC DERMATITIS” to go the AD intercellular communication map
1.5. In this particular application, we want to check possible pathways in KC that could attenuate the effect of dupilumab. So, go to “Submaps” and then click “>” associated with the submap “Keratinocyte”.
1.6. A new tab (“Keratinocyte”) showing the intracellular pathways of KC appears. Click the magnifier icon until the point you are able to identify the proteins.
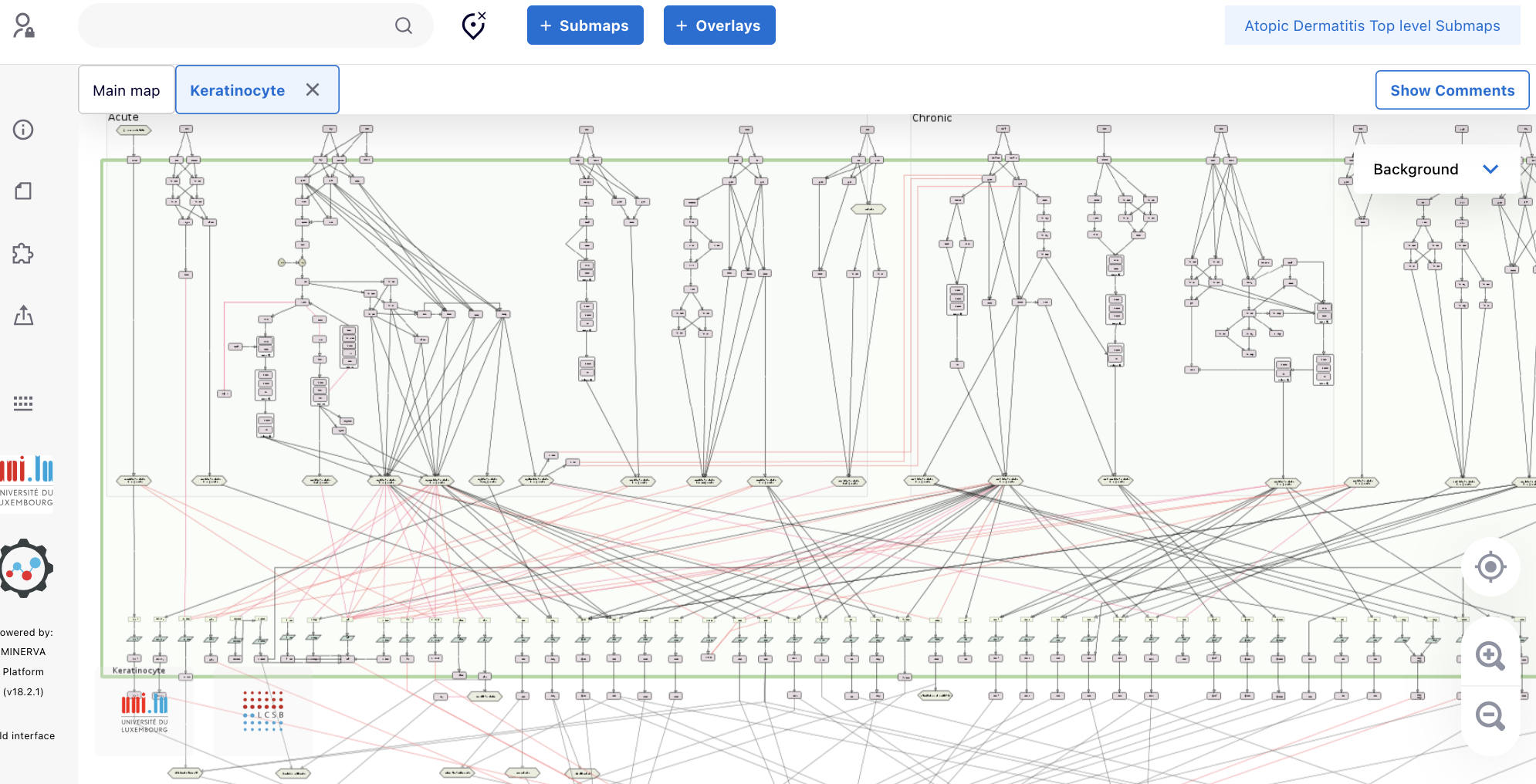
2. Exploring the structure of the map
2.1 To identify pathways couteracting dupilumab’s effects, first we have to find all secreted proteins that are positively regulated by the IL4/IL13 signaling (in this case, either the phenotype “IL4/IL13 induced regulation of gene expression” or “IL4 induced regulation of gene expression”). To this end, pay attention only to the black connections to the proteins and click them. After clicking, you can see in the left side of the map a panel showing information about the target gene/protein, including the source of the selected connection (at the botton). After checking all proteins, we can see that TNFAIP6, IL25, CCL26, CXCL8, TSLP, TNC, VCAM1 and POSTN are positively regulated by the IL4/IL13 signaling pathway.

2.2. Among these proteins, check the ones that are regulated by at least one additional protein or signaling pathway, i.e. one additional black connection pointing to them. Only by simple visual inspection, we can observe that CXCL8, TSLP and TNC are also regulated by other protein and/or signaling pathways.
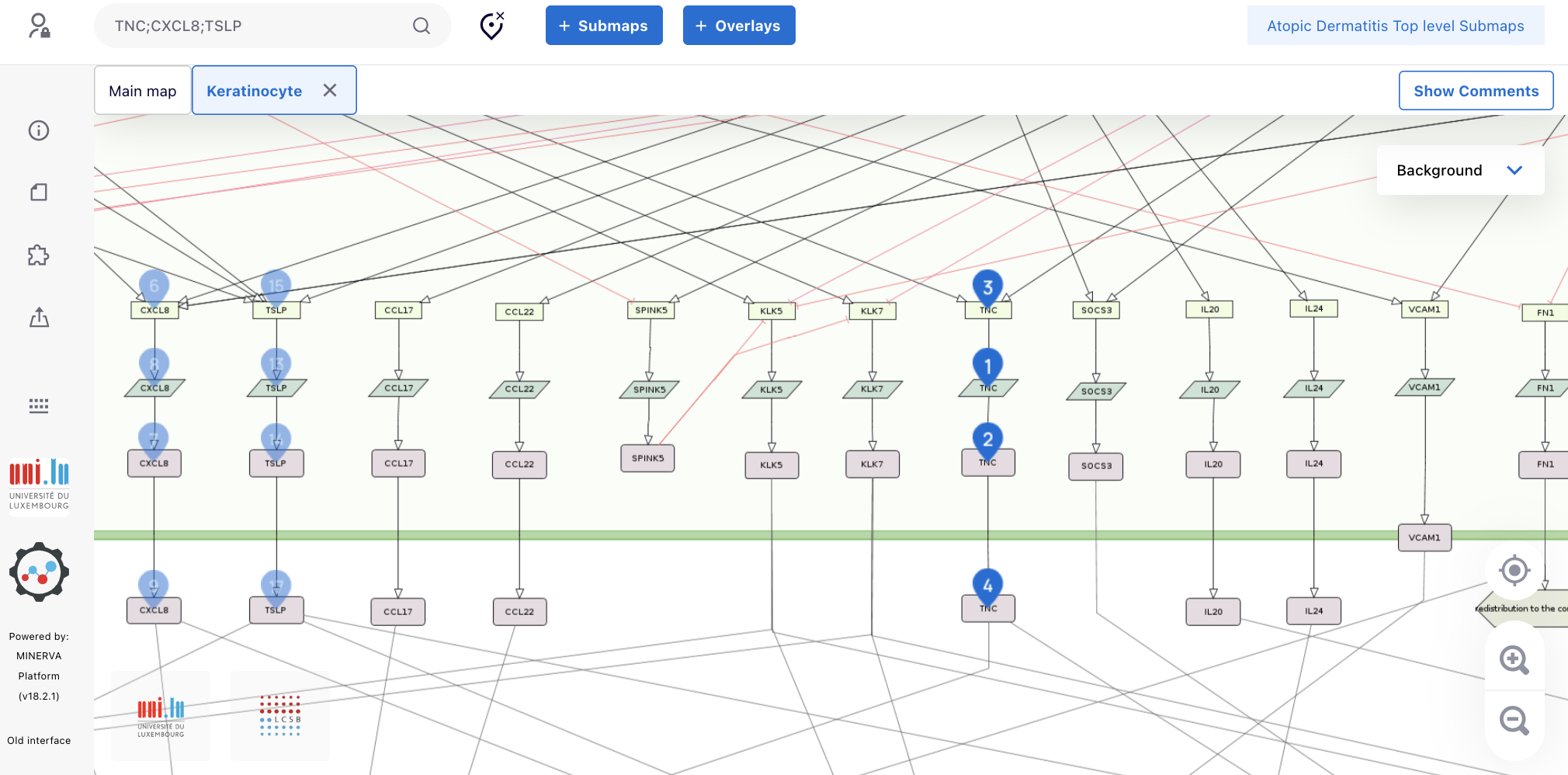
2.3. To find out which signlaling pathways also regulate CXCL8, TSLP and TNC, click the additional connections and, after clicking, look at the panel in the lefs side of the map showing information about the target gene/protein, including the source of the selected connection (at the botton). We see that, in addition to IL13/IL4 signaling pathway, CXCL8 is also regulated by IL26 and IFNG signaling pathways, TSLP is also regulated by IL33 and IL26 signaling pathways and TNC is also regulated by INFG signaling pathway.
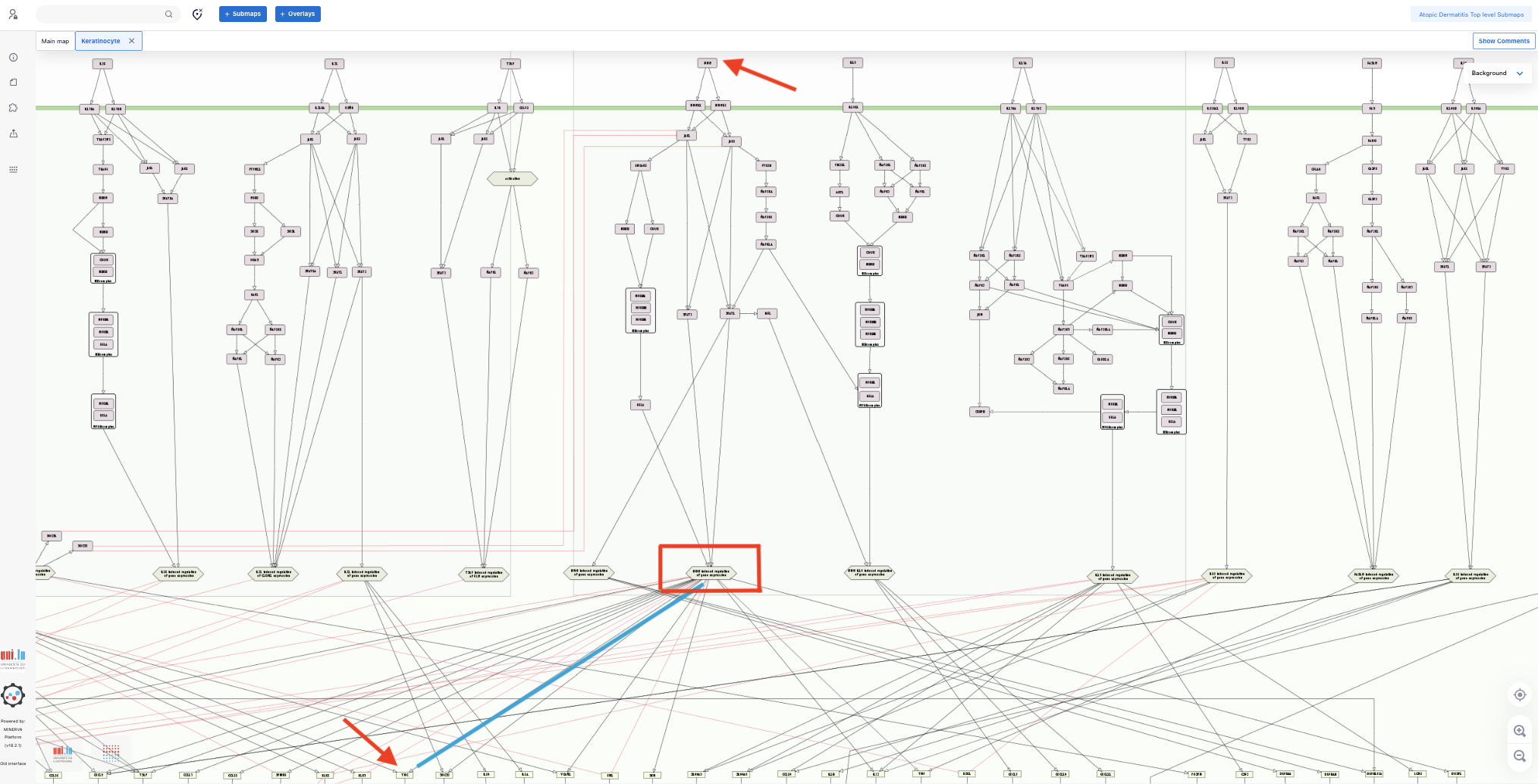
3. Hypothesis
IL33, IL26 and IFNG signaling pathways in KC partially mitigates the anti-inflammatory effect of dupilumab.