
Step-by-step guide to reproduce the application “SNPs in upstream apoptosis regulators in psoriatic KCs drive their resistance to cytokine-induced apoptosis”
1. Obtaining the genetic variant data
1.1. Collect genes harbouring variants (SNPs) associated with PsO from the Open Targets database. First, access the Open Targets Platform’s home page.
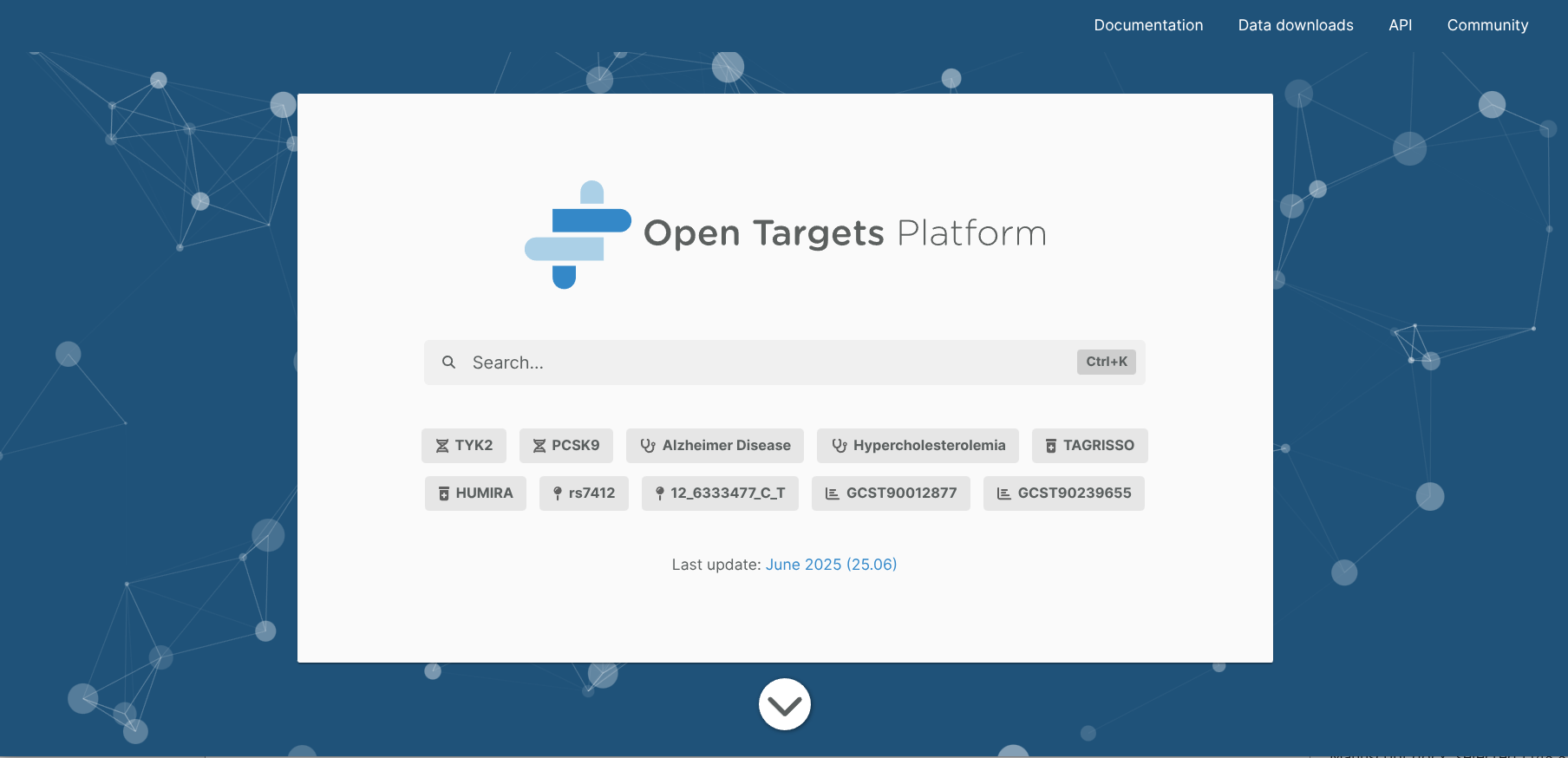
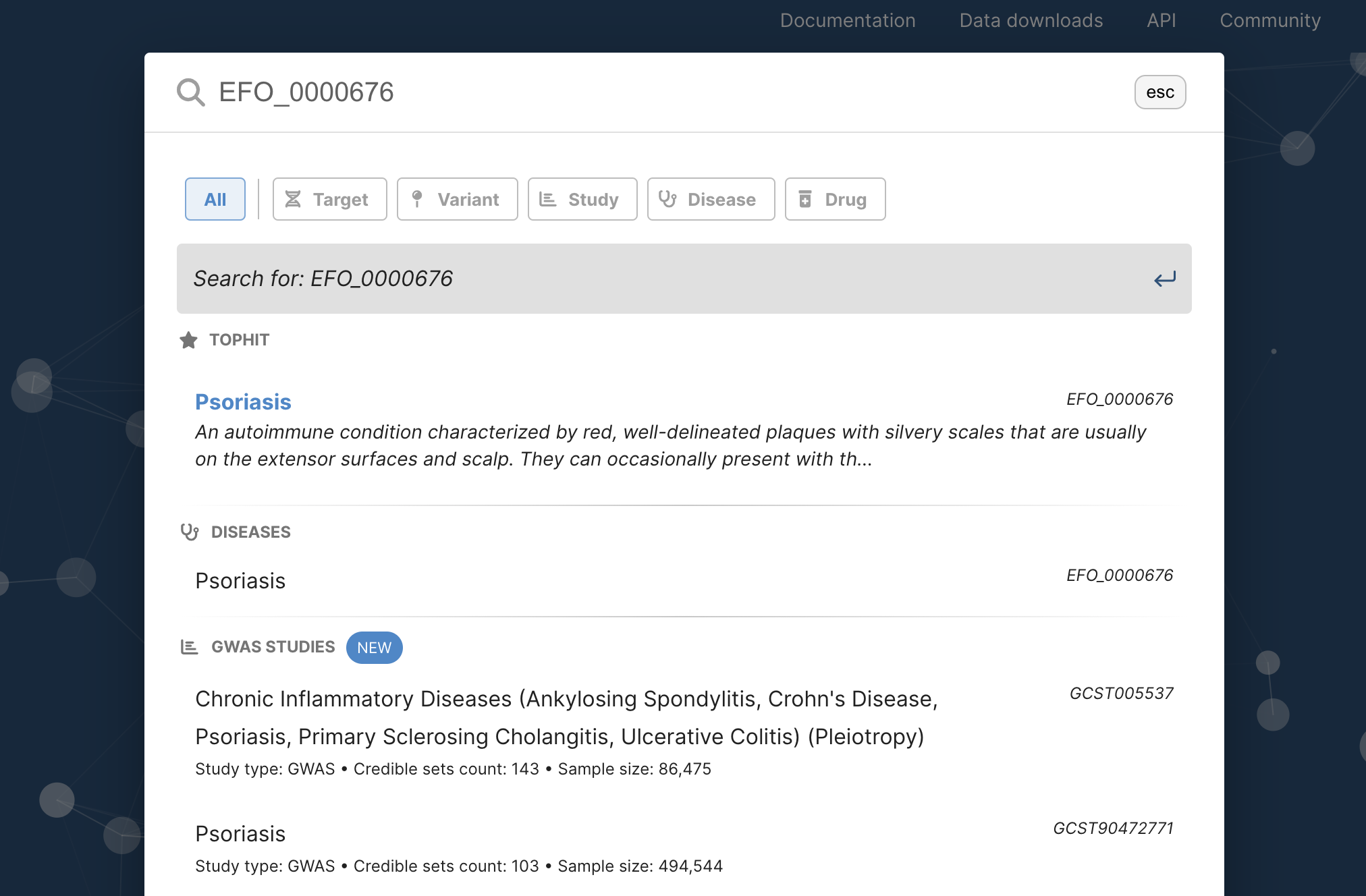
1.2. Then, use the Experimental Factor Ontology (EFO) identifier of PsO (EFO_0000676) as a query. As soon as you finish typing, “Psoriasis” appears on the screen. Click it.
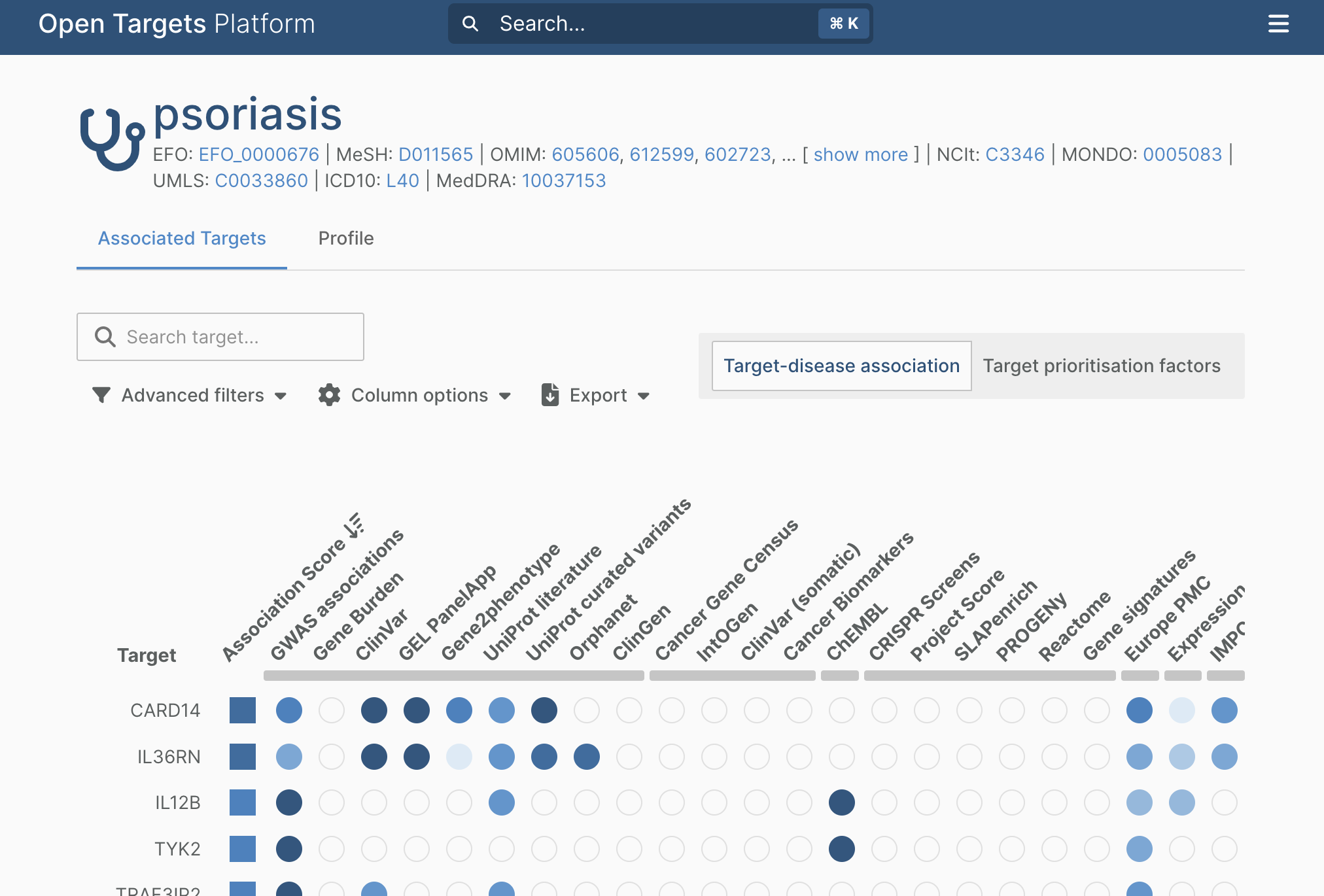
1.3. A page containing the retrieved results appears. Go to “Columns options” and selected the following sources of genetic data: GWAS associations, ClinVar and Uniprot Curated Variants
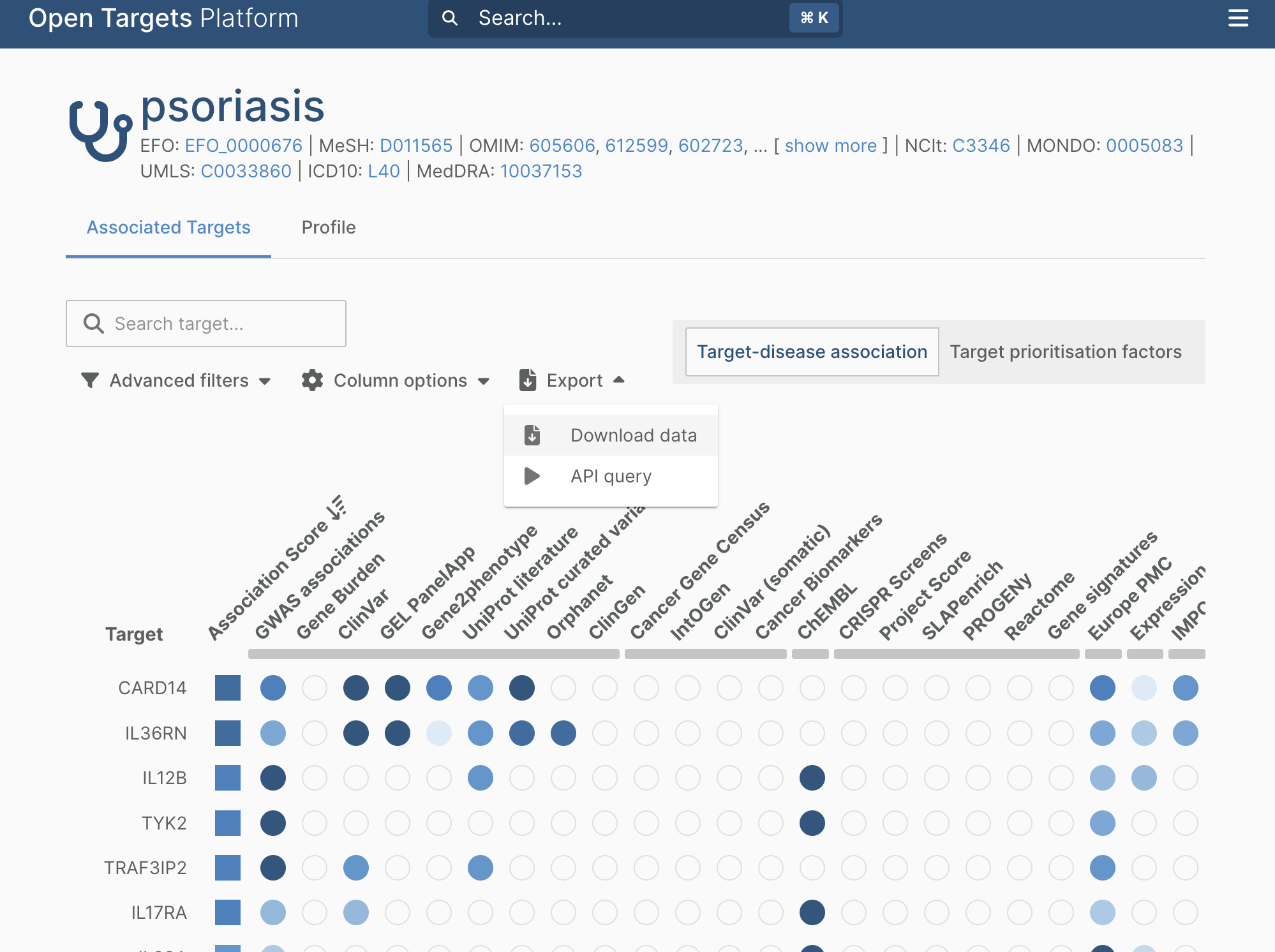
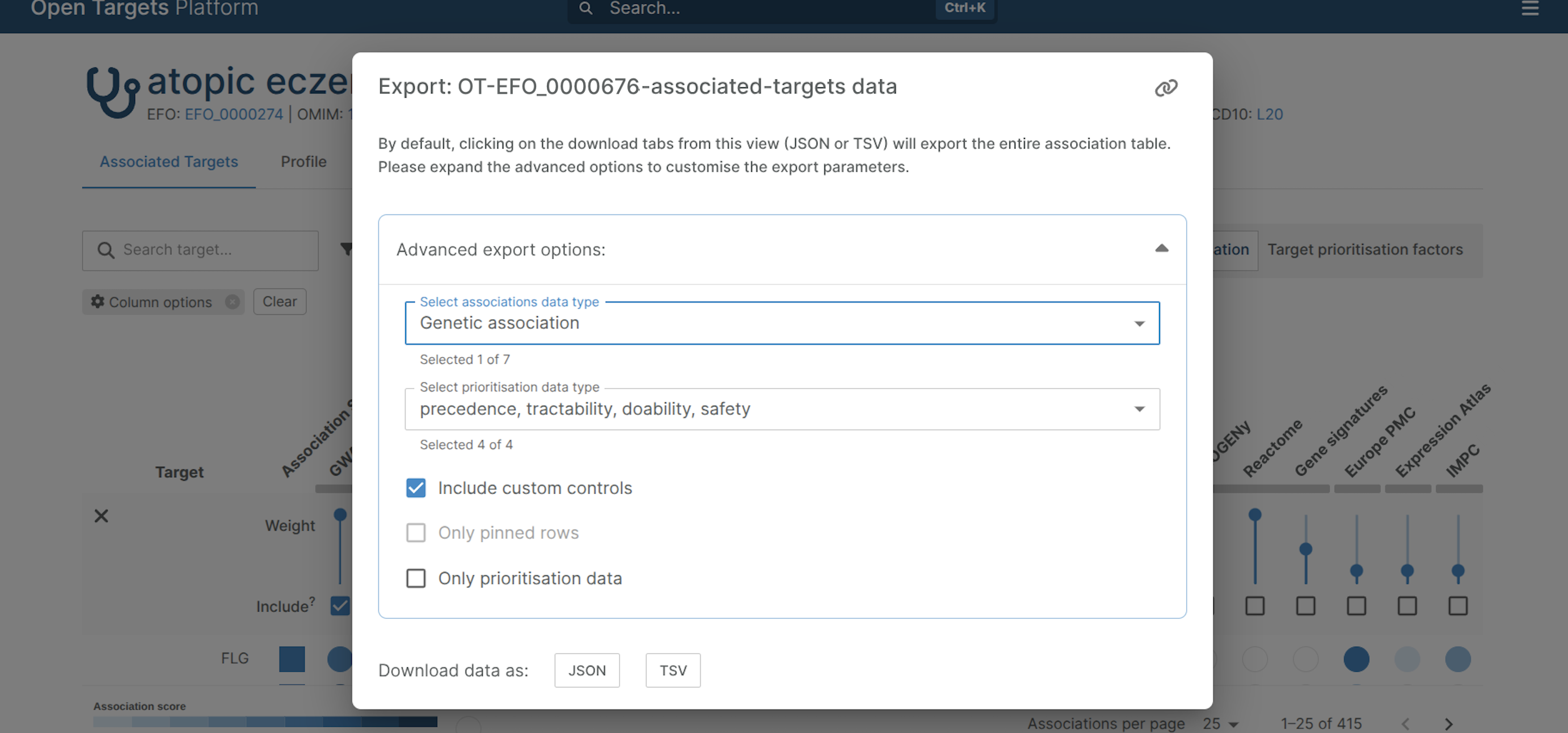
1.4. Click “Export” and then “Download data”
1.5. A new window appears. Click “Advance Expert Options” and, in the field “Select association data type”, select only “Genetic association”
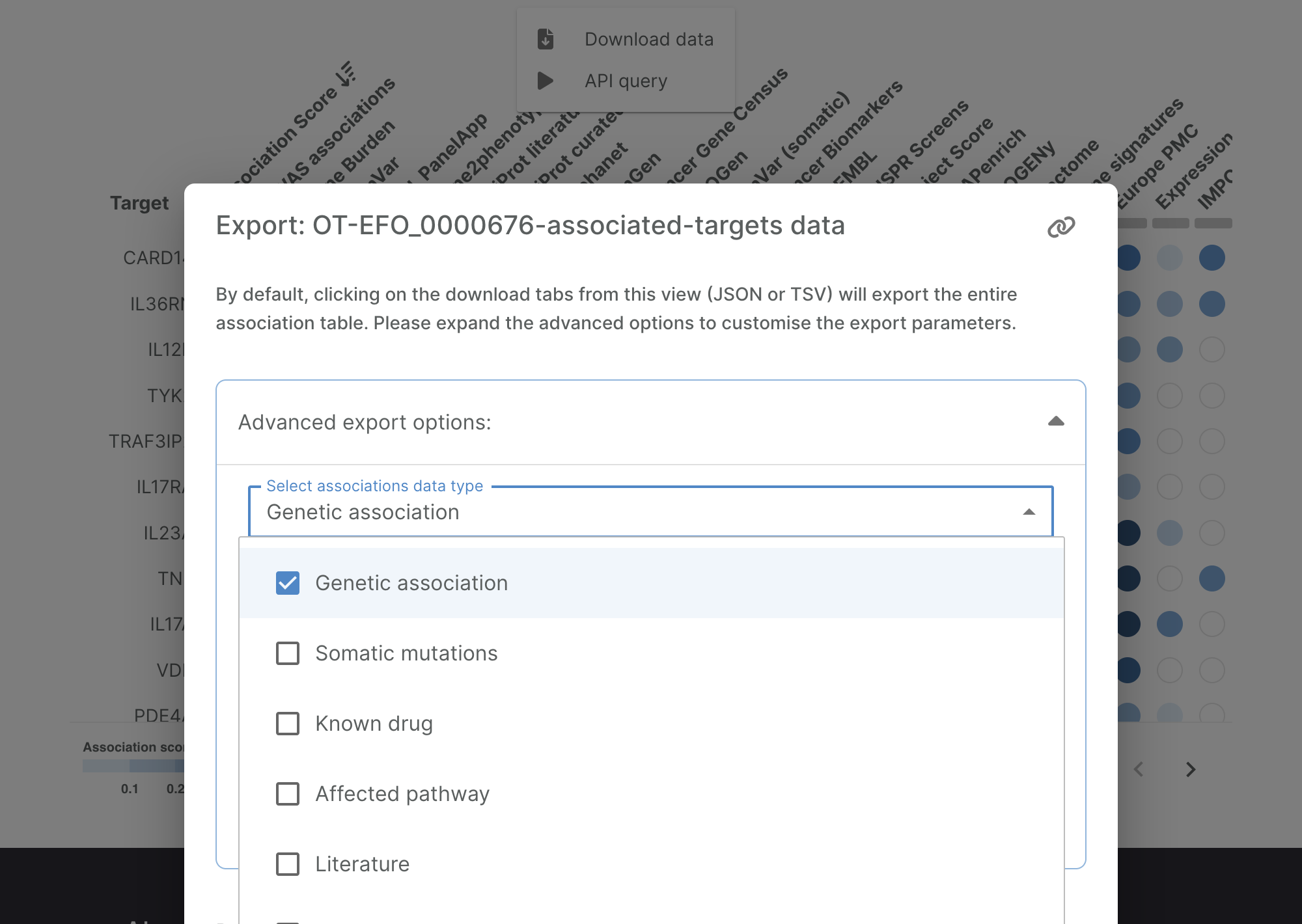
1.6. Finally, click “TSV” to download the file containing the PsO-associated genes in a TSV format. Make sure that the option “Include custom controls” is selected.
2. Preparing the data-containing file for integration
2.1. Open the downloaded TSV file in Excel or similar software. Copy the contents of column 1 (symbol) and paste them into a new spreadsheet.
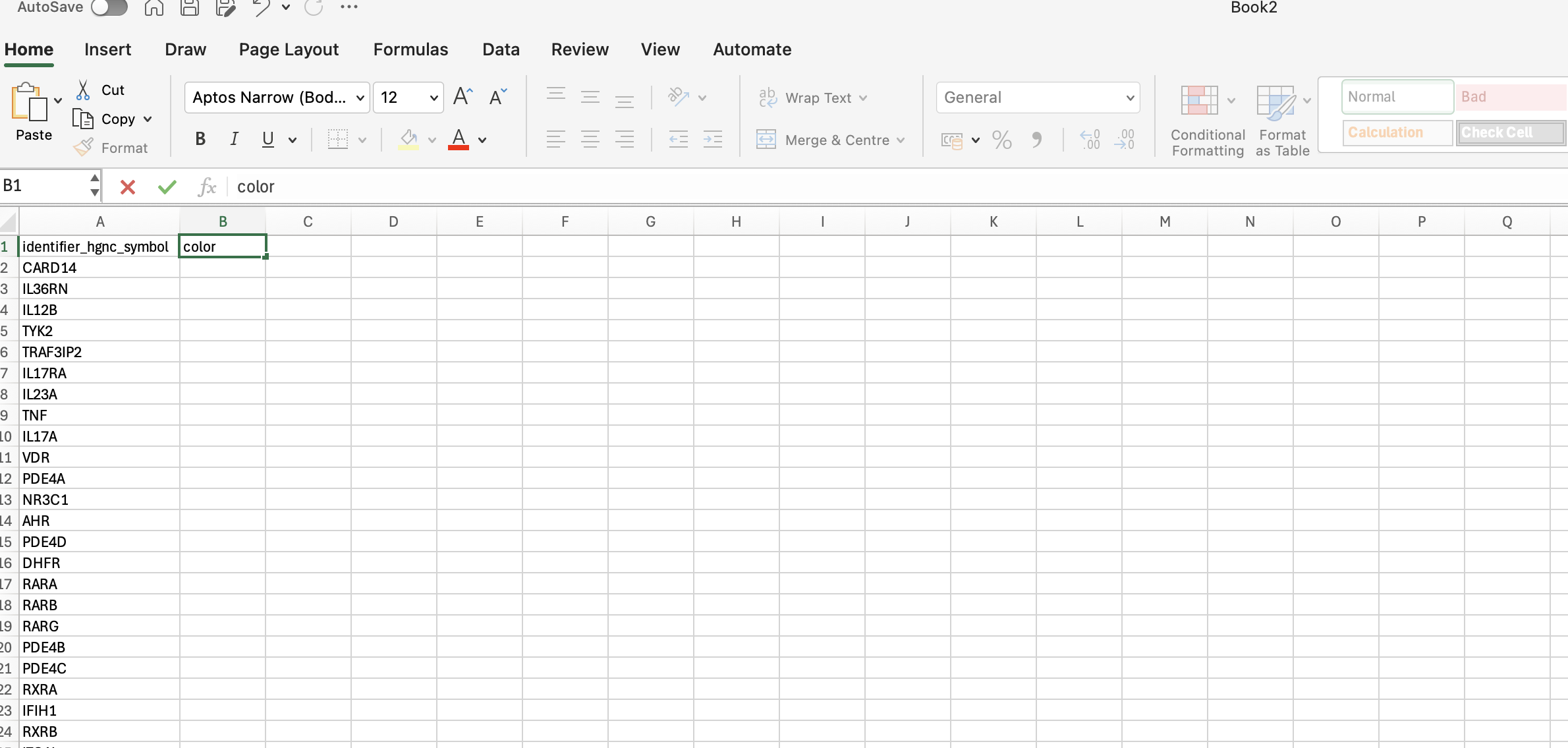
2.2. Create a header: name the column 1 as “identifier_hgnc_symbol” and for column 2 as “color”. The list of genes should start in row 2.
2.3. Fill rows in column “color” with a hex color code for your color of interest. Suggestion: #40652e (green). Save this file as “PsO_genetic1.txt”
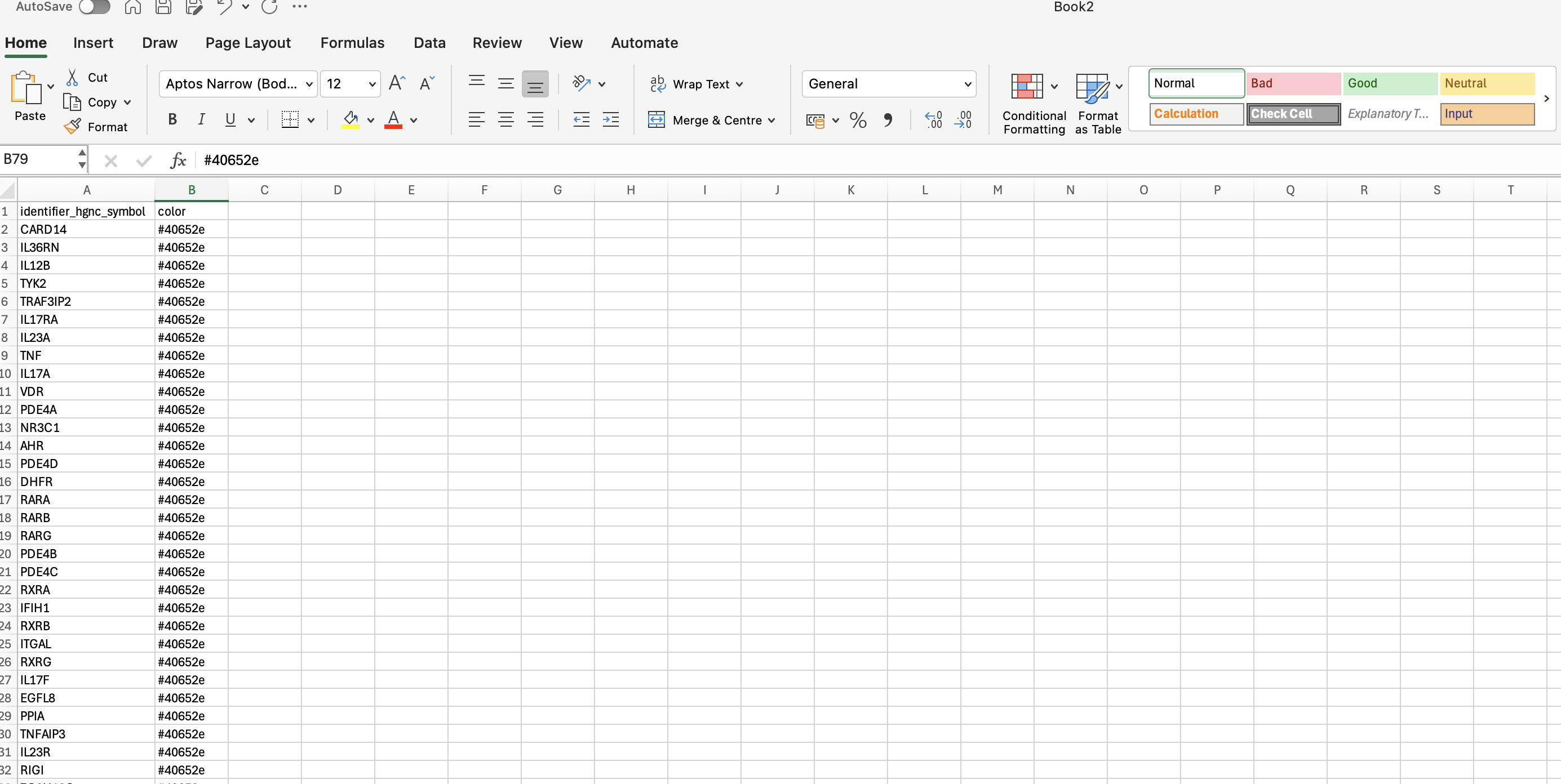
3. Access and log in to the map
3.1. Access the ISD map at the entry level via the link (https://imi-biomap.elixir-luxembourg.org/). In the (map), click the login icon in the left upper side of the screen. This is required to integrate data into the map.
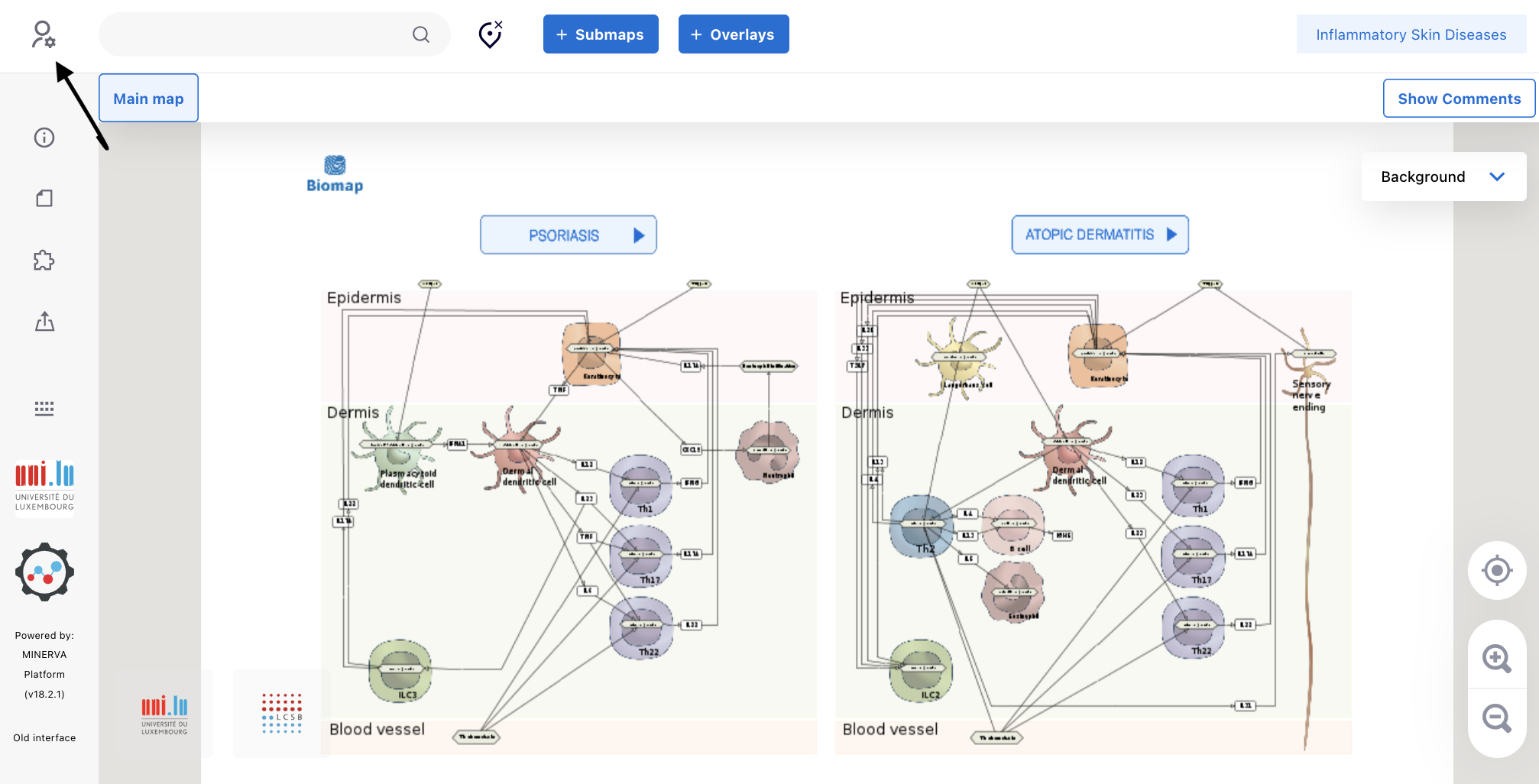
3.2. Log in to the map preferentially by using your ORCID.
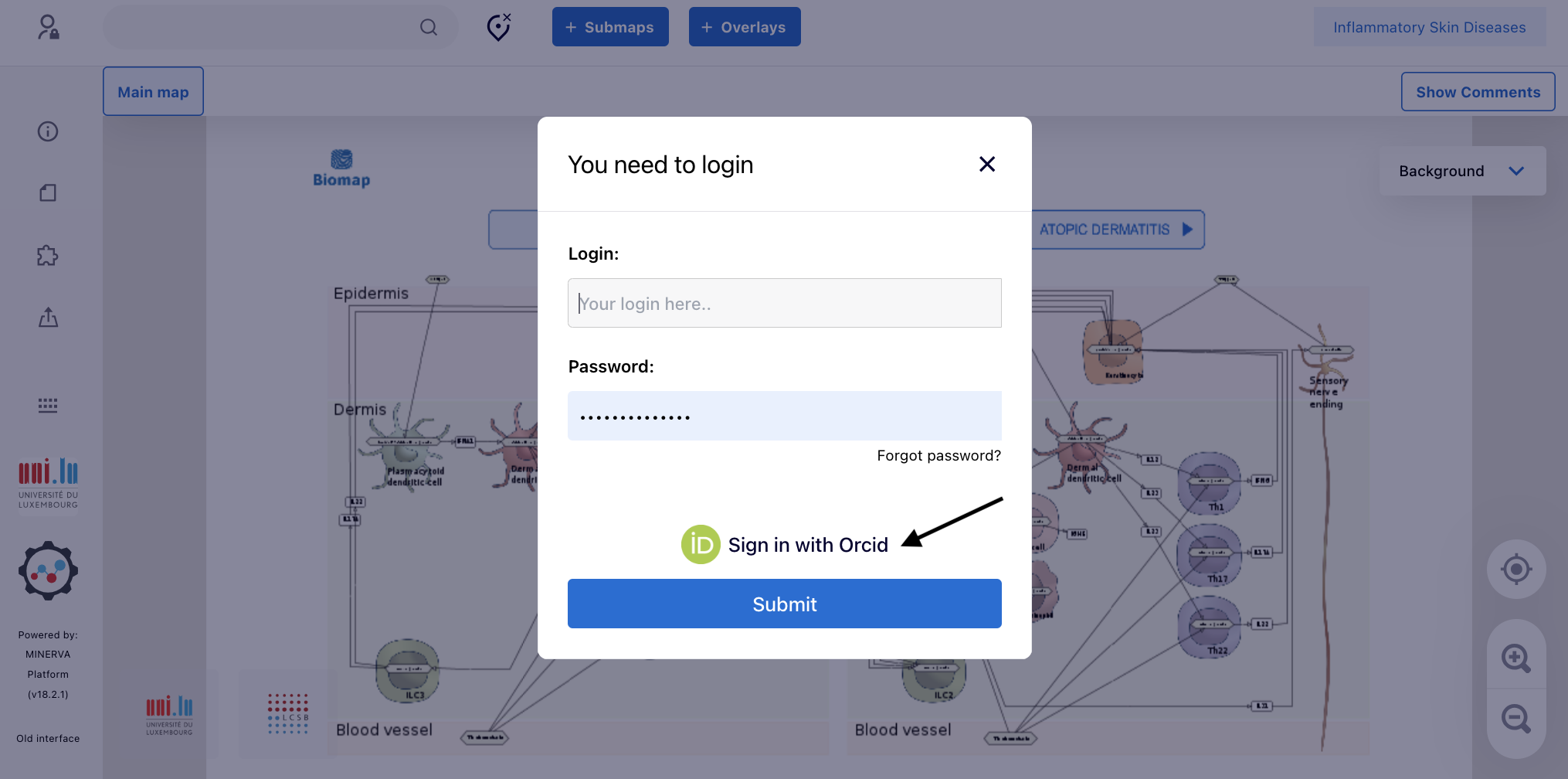
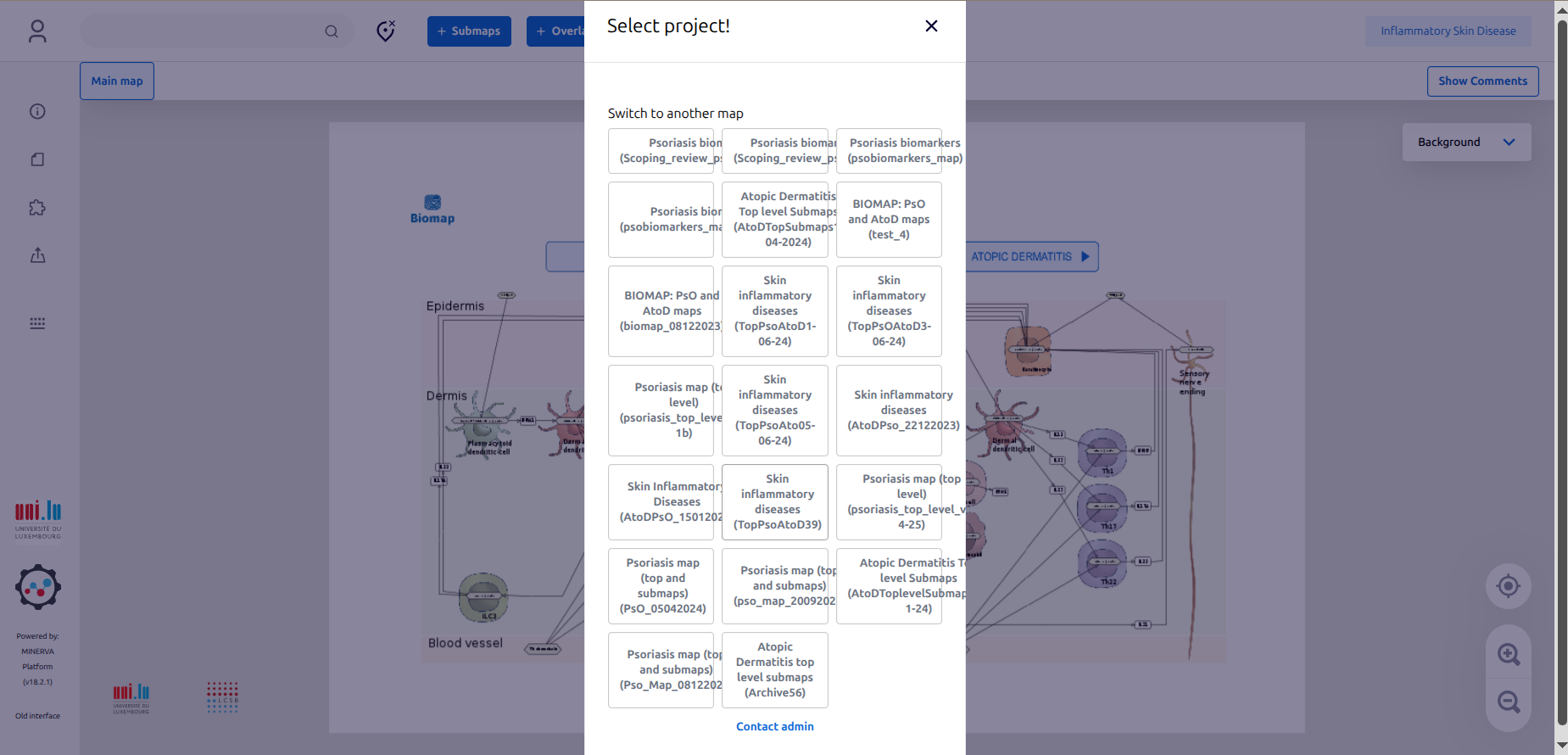
3.3. Once connected, just ignore the window “Select project” by clicking “X”.
3.4. Click the button “PSORIASIS” to go to PsO intercellular communication map
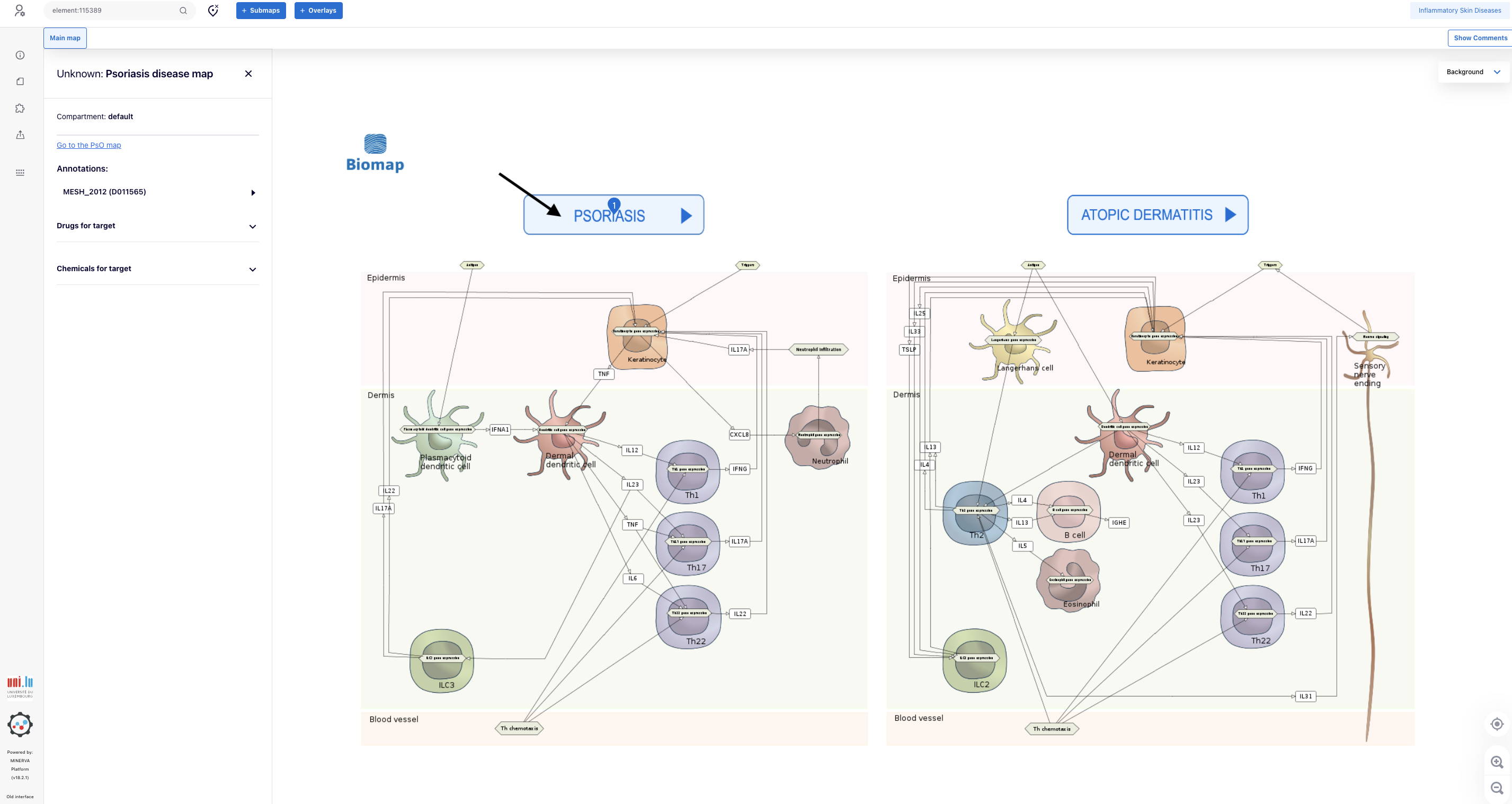
4. Create the overlay for integration
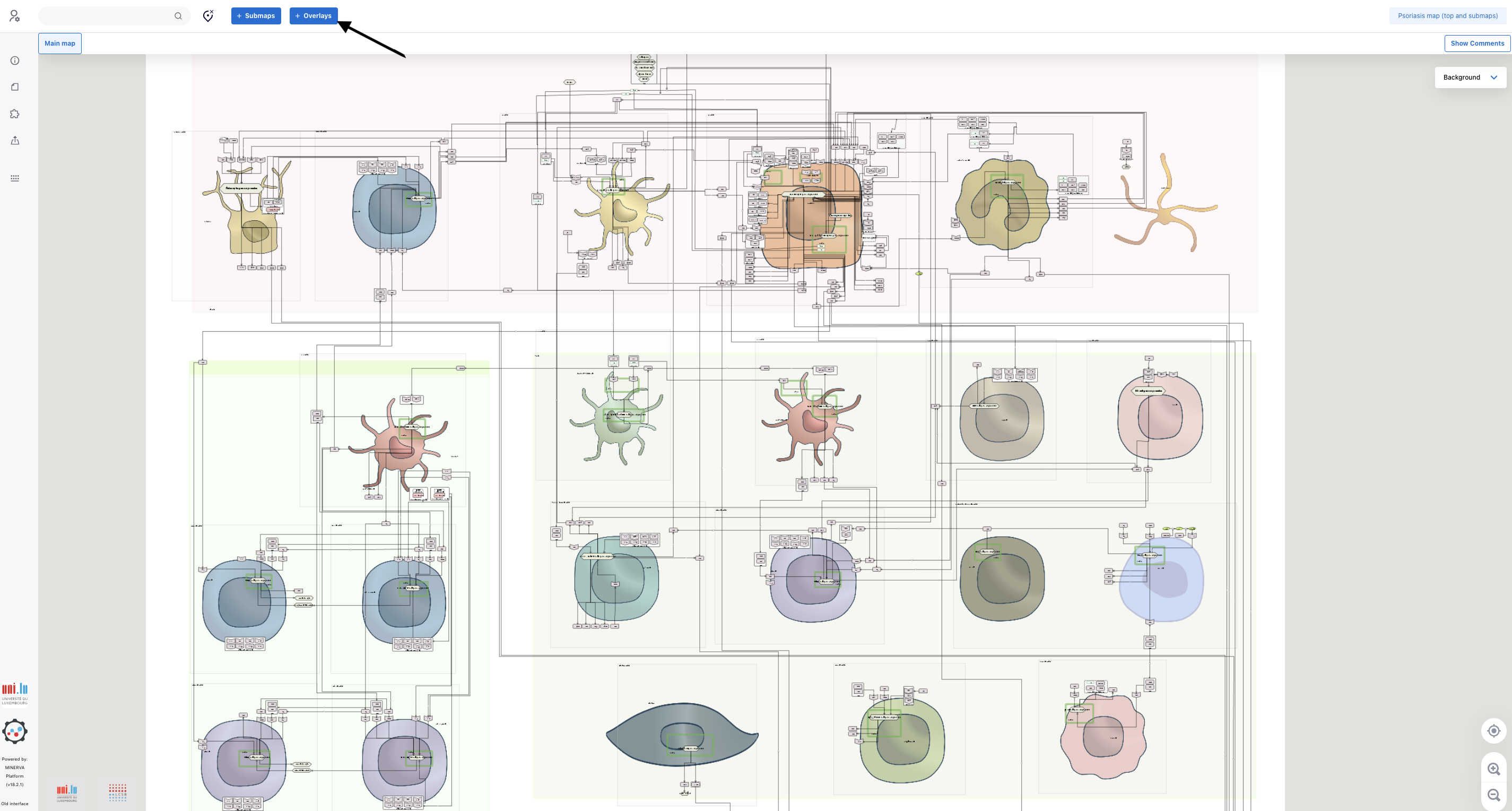
4.1. The integration of data into the map is done via overlay creation. For this purpose, click the button “+ Overlays” above the intercellular communication map
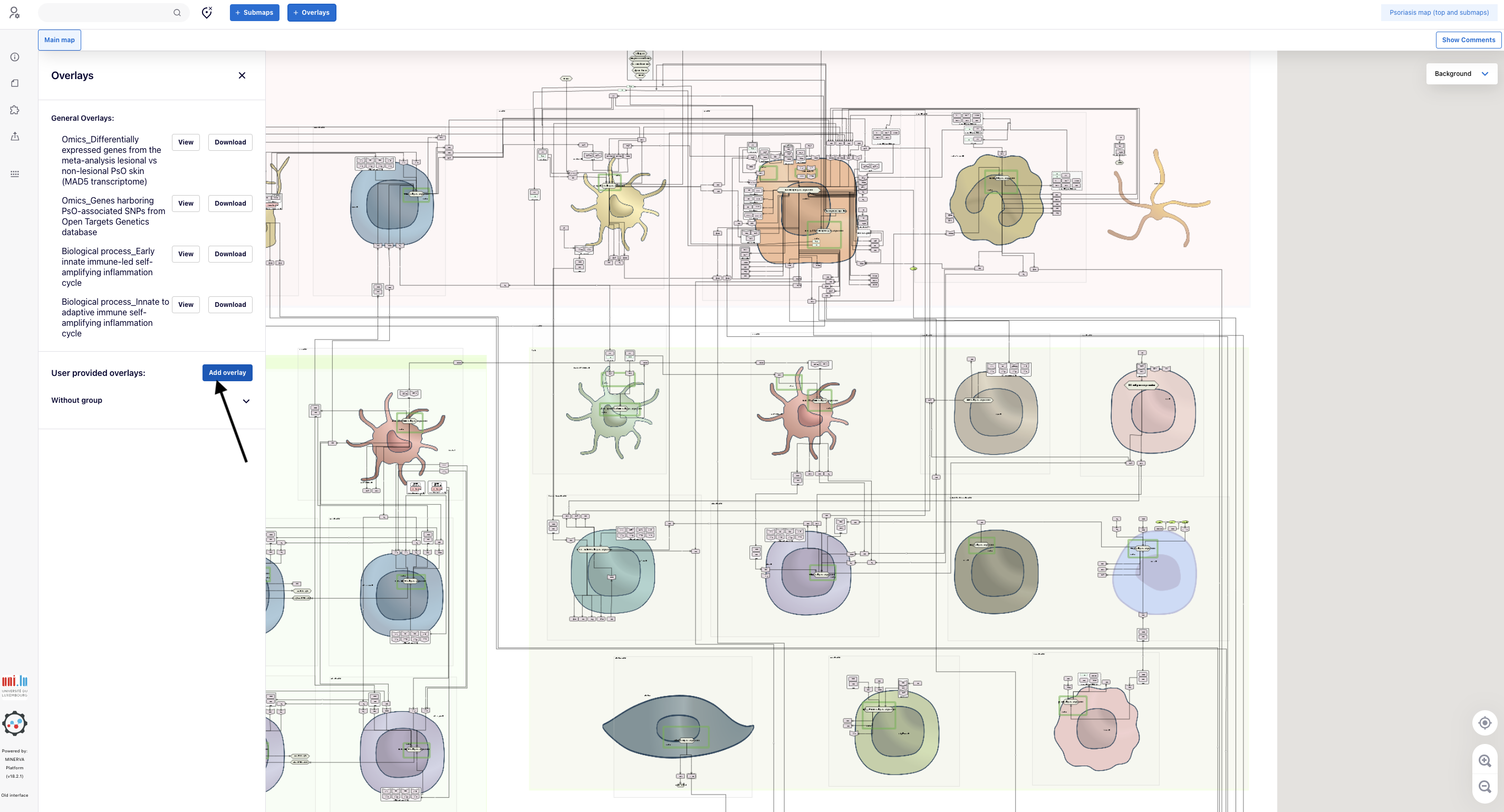
4.2. When the panel “Overlays” opens in the left part of the screen, go straight to the bottom and click “Add overlay”
4.3. In the panel “Add overlay”, click “browse” to upload the file “PsO_genetic1.tsv”.
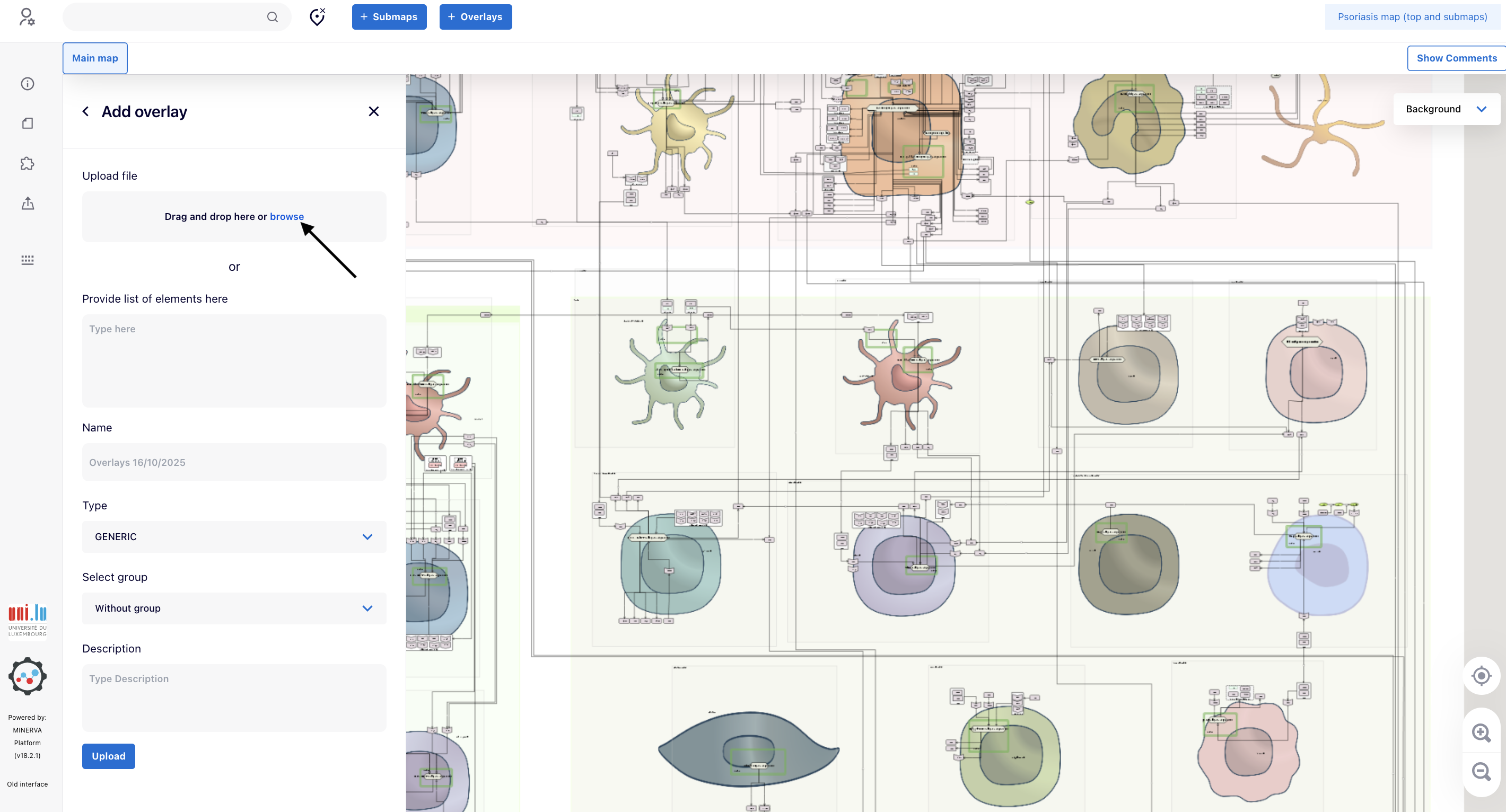
4.4. When you observe “PsO_genetic1.tsv” instead of “browse”, provide a name in the field “Name” (e.g., “PsO genetic”). Go to the bottom and click “Upload”
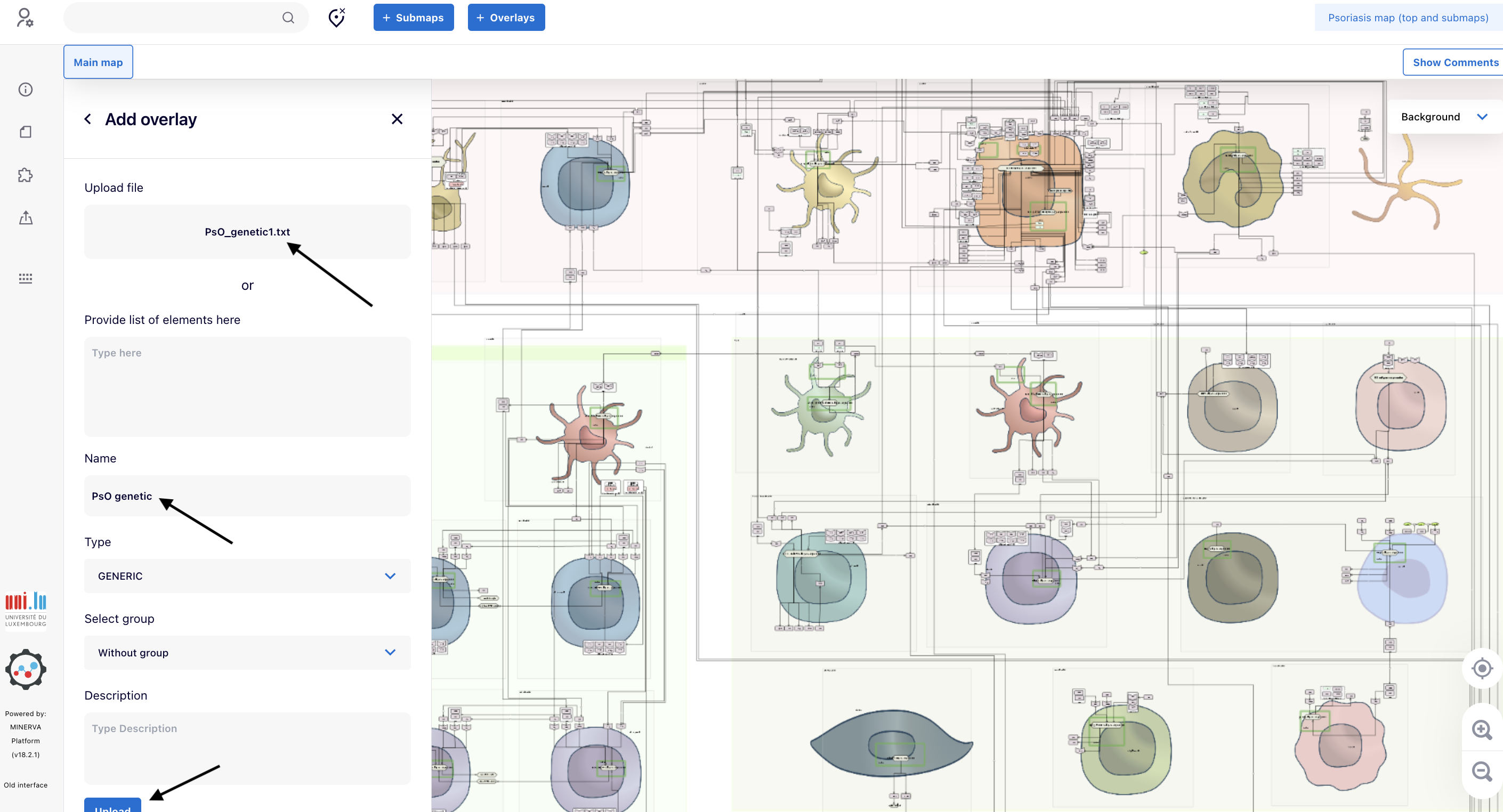
4.5. When you observe the warning “User overlay added successfully”, click “<” close to “Add overlays” to go back to the “Overlay” panel and initiate the explorations as shown in next steps.
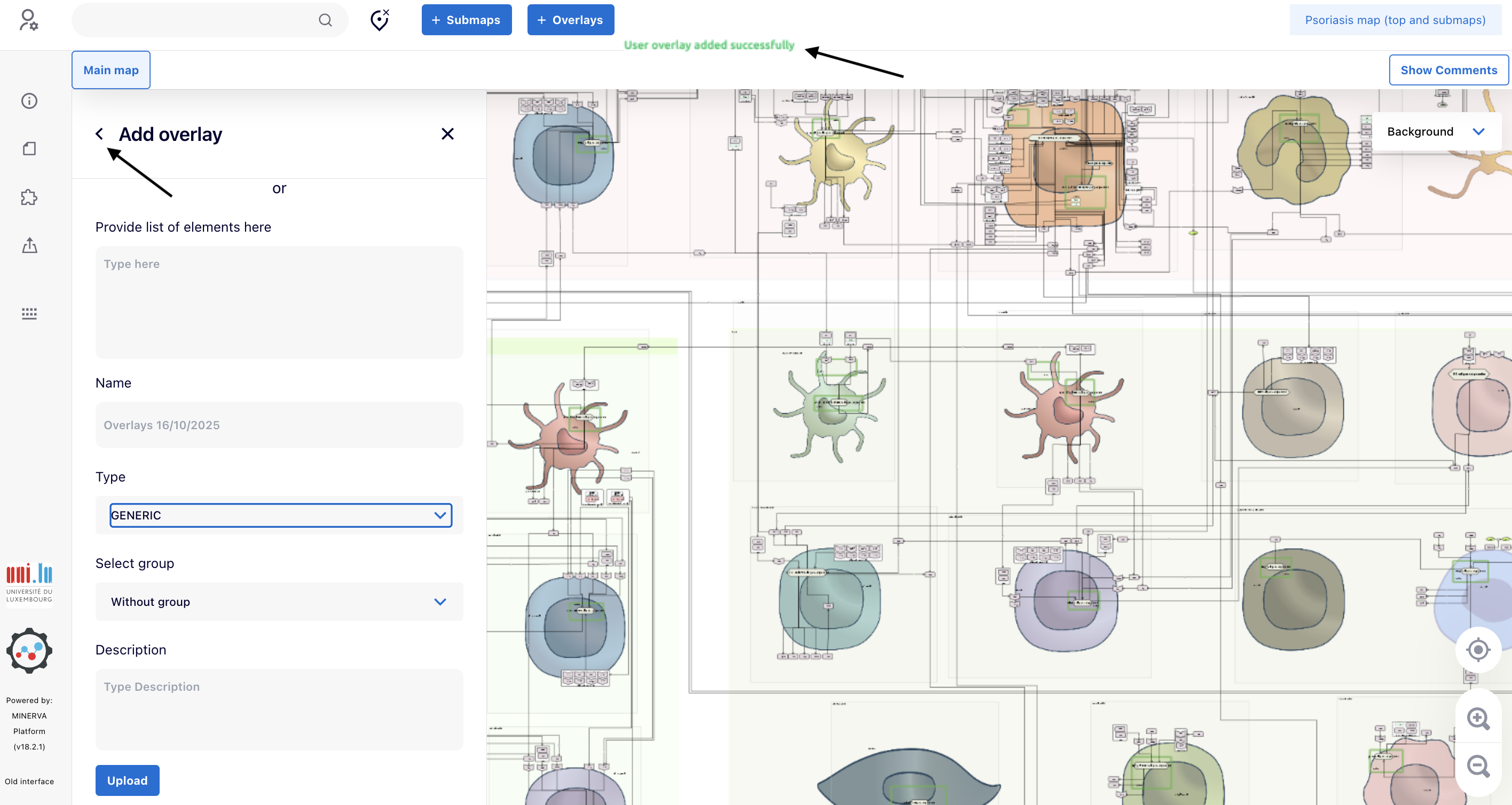
5. Integrating and exploring the data into the map
5.1. Access the genetic data, i.e., the overlay “Pso genetic”, via the panel “Overlays”. Go to the bottom and click “Without group”. The overlay “PsO genetic” appears.
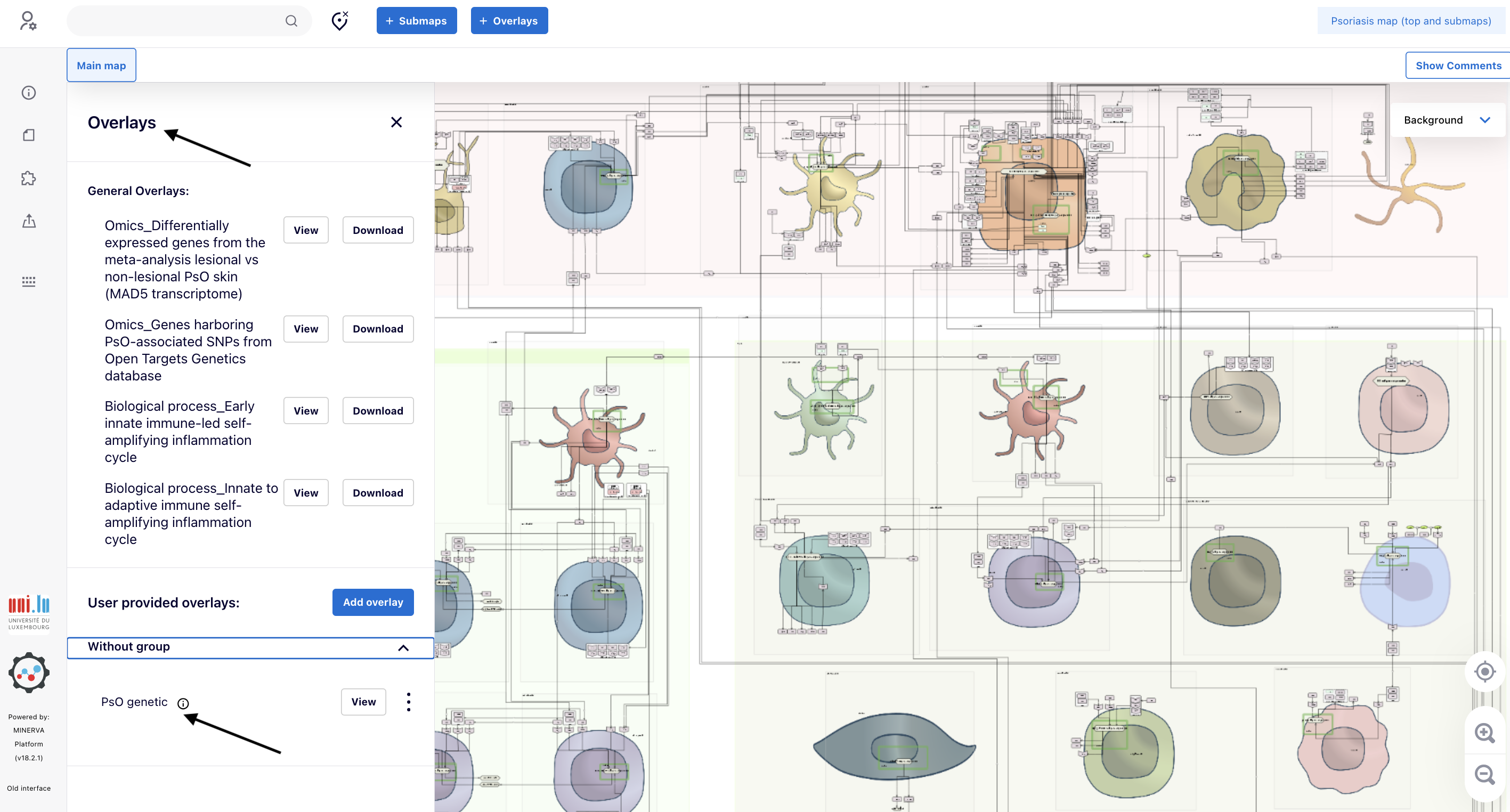
5.2. Click “View” and the proteins matching the genes harboring PsO-associated SNPs will be painted in green (link to the map).
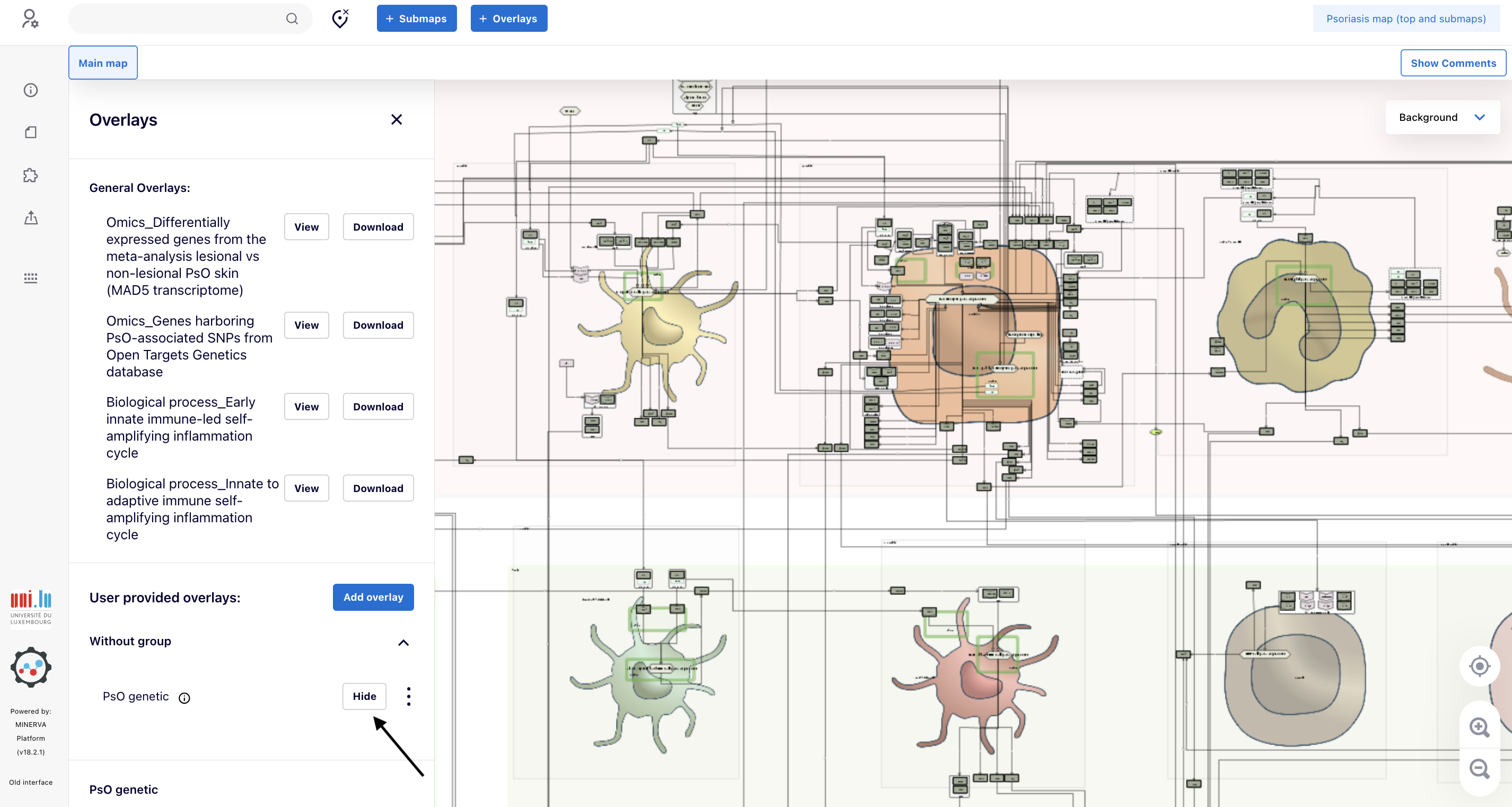
5.3. In this particular application, we want to check the intracellular pathways in keratinocytes. Go to “Submaps” and then click “>” associated with the submap “Keratinocyte”
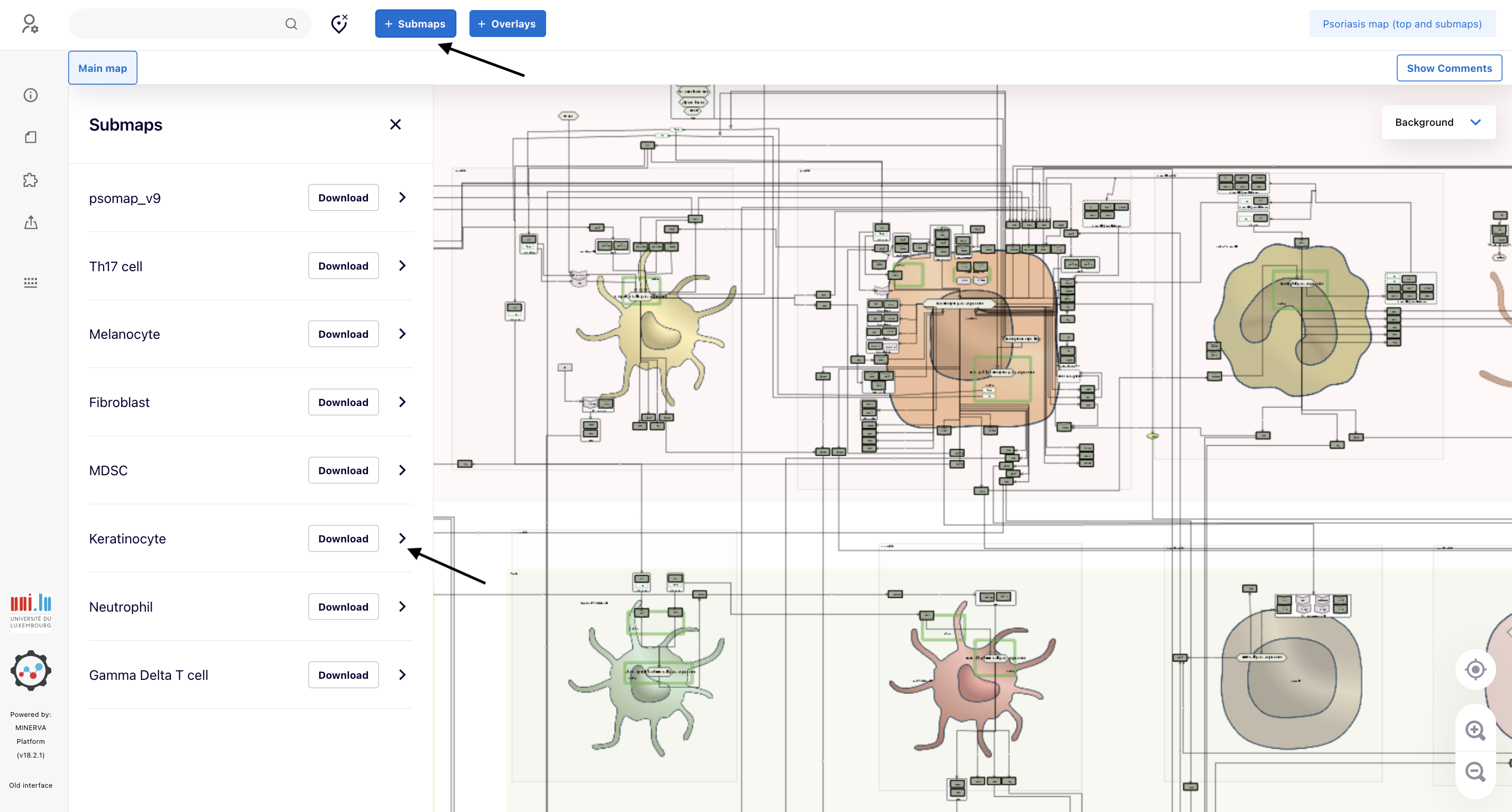
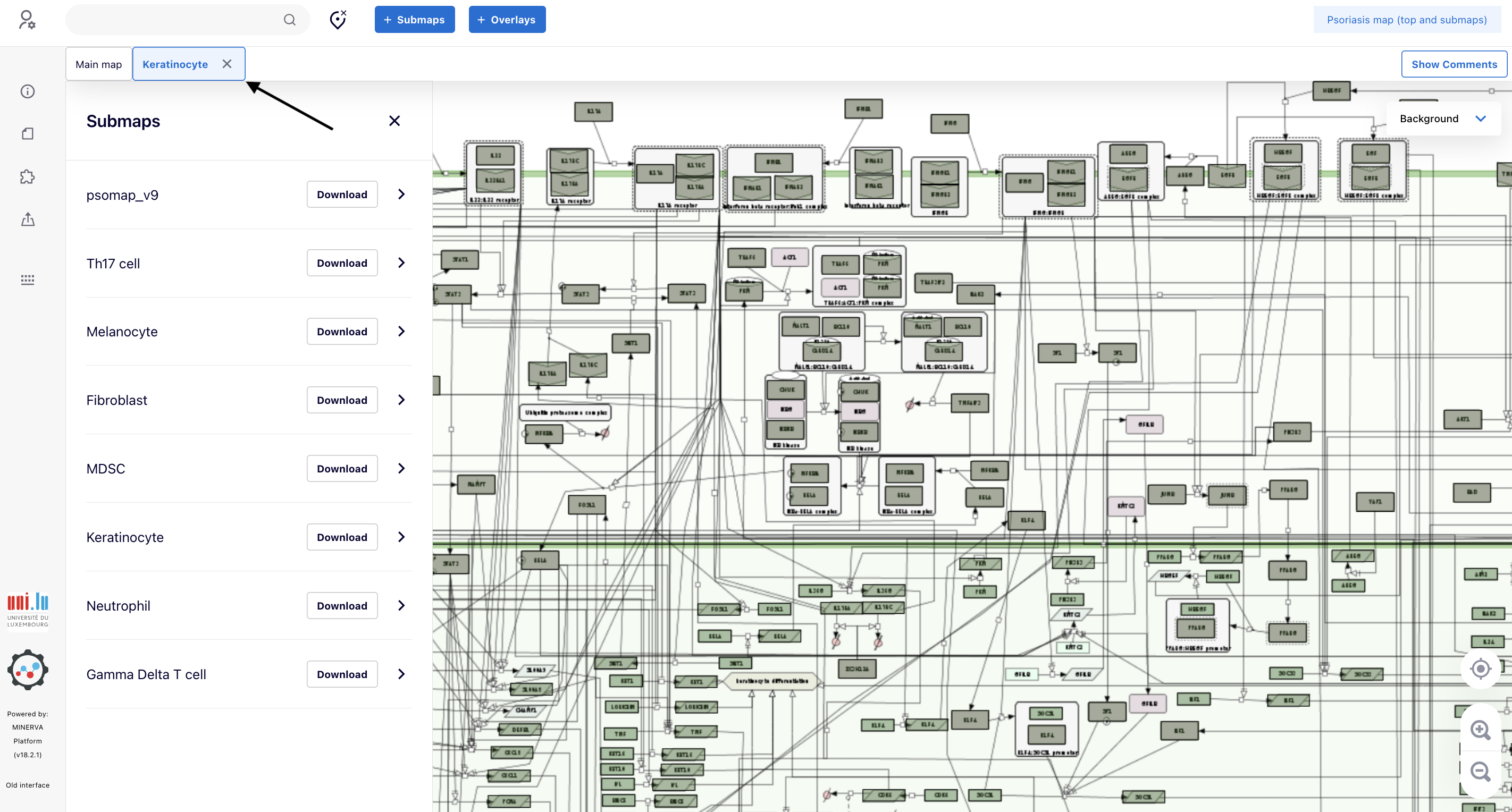
5.4. A new tab (“Keratinocyte”) showing the intracellular pathways of Keratinocyte appears. It is possible to see some proteins painted in green. To take a further look at them, click the magnifier icon until the point you are able to read the gene symbols in the elements (link to the map).
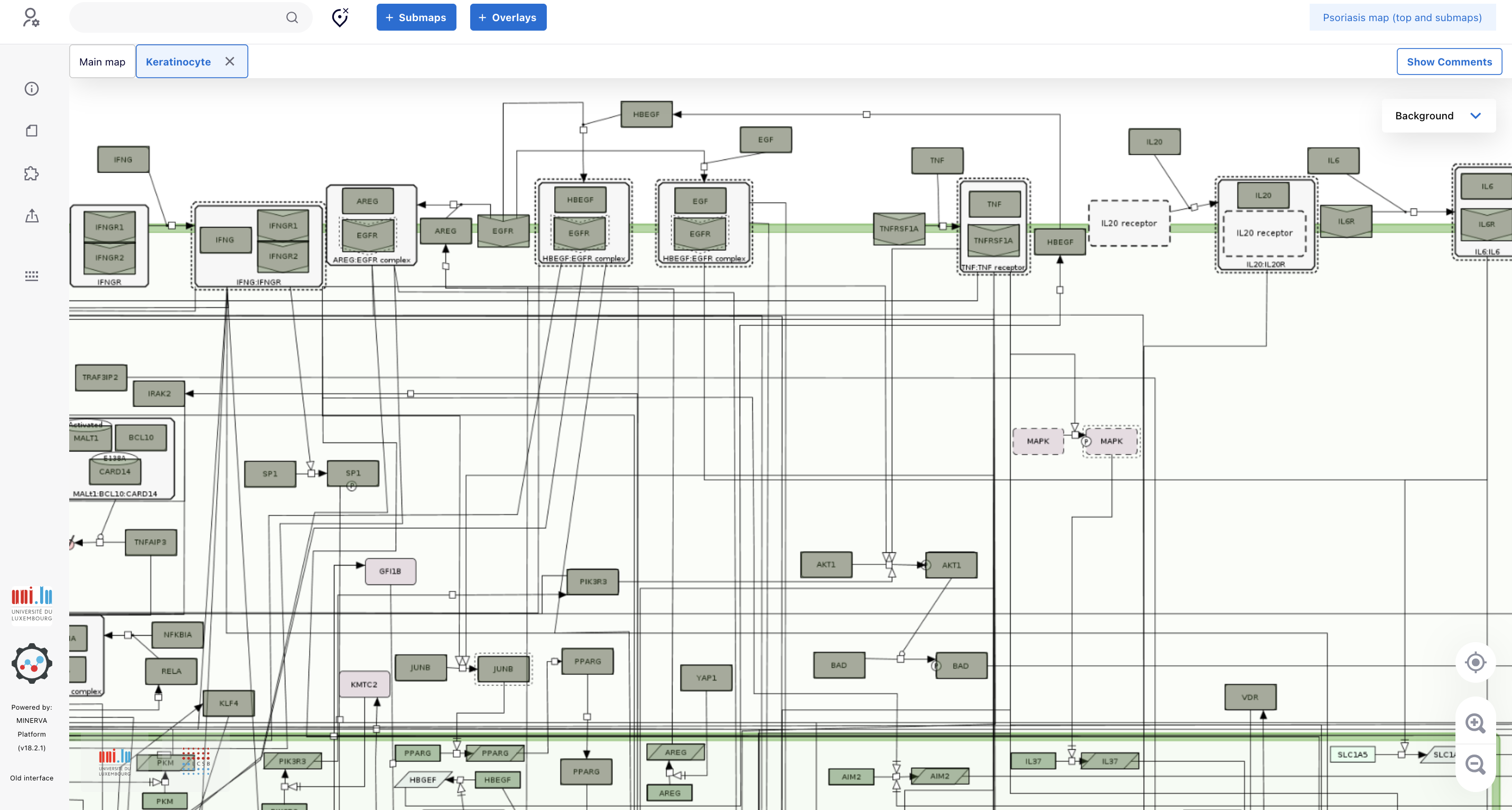
5.5. By navigating the map at the zoom level of your convenience, you can see that there are proteins coded by PsO-associated genes: IFNG, INFGR2, TNFRSF1A, ESRRA, IRF1 and SOCS1.
5.6. There is a direct pathway where all proteins are encoded by PsO-associated genes and the underlying SNPs could favour the inhibition of apoptosis in KC.
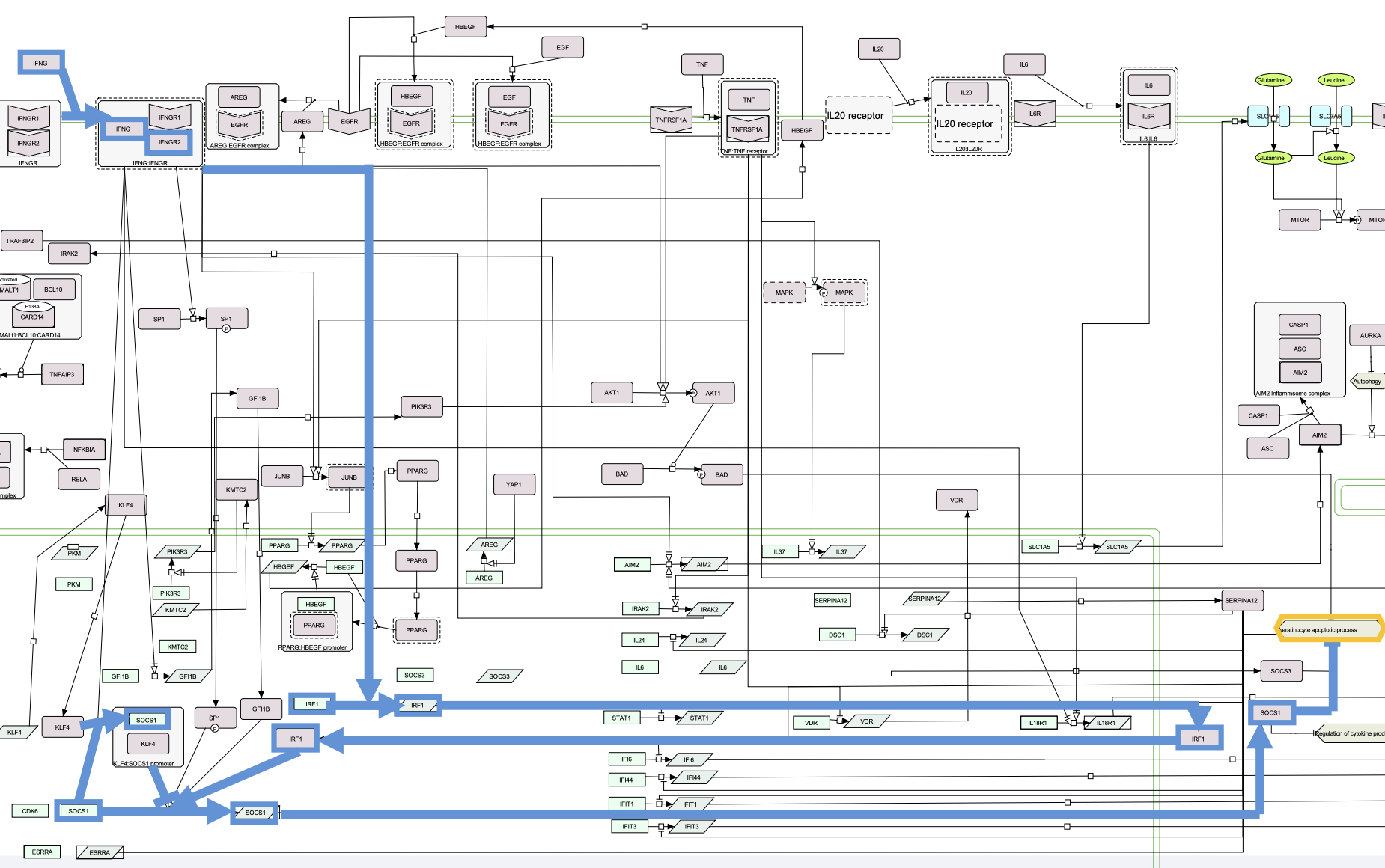
6. Hypothesis
There are at least six proteins encoded by PsO-associated genes in this pathway: IFNG, INFGR2, TNFRSF1A, ESRRA, IRF1 and SOCS1. It is the most prominent pathway triggered by IFNG via IFNGR2 and IRF1 culminating in the expression of SOCS proteins.